Introduction
Materials and Methods
Rhizosphere Soil Collection
Culture Media
Soil Microbial Isolation and Purification
Physiological and Biochemical Identification
DNA Extraction and Illumina Sequencing
Sequence Alignment and Cluster Analysis
Results
Soil Micro-Ecology
Colony Characteristics and Microscopic Morphology of Bacteria, Actinomycetes, and Fungi in the Soil of Continuously Cropped Pepper
Physiological and Biochemical Properties of Bacteria and Actinomycetes in the Soil of Continuously Cropped Pepper
Sequencing and Cluster Analysis of Bacteria, Actinomycetes, and Fungi in the Soil of Continuously Cropped Pepper
Discussion
Introduction
Pepper blight caused by Phytophthora capsici devastates pepper production and can spread quickly in a short time (Kim et al., 2002; Zhang et al., 2008; Barchenger et al., 2018). Since its isolation from pepper for the first time in 1918 in New Mexico, United States (Leonian, 1922), nearly 80 pathogenic species have been found residing in soil in various states such as mycelium, sporangium, chlamydospores, and spores (Judelson and Blanco, 2005). Therefore, P. capsici not only has the ability to resist rhizosphere stresses, but can also easily activate to infect pepper. New sporangium and zoospores on pepper spread in the soil by rain or through irrigation water, which could lead to a large infestation in the area at the seedling stage. It may also spread through the air, infecting the plant stem, leaves, flowers, and fruits (Silvar et al., 2006). Recently, large production losses caused by pepper blight have been documented in many countries with particularly devastating effects in the United States, Brazil, India, Mexico, Russia, South Korea, Japan, Bulgaria, France, Italy, Argentina, and Pakistan (Hao et al., 2007; Barchenger et al., 2018; Nawaz et al., 2018; Reyes et al., 2019; Reyes et al., 2019). In China, pepper blight was first discovered in Jiangsu province in the 1950s; more than 10 provinces are reporting serious pepper blight infections, especially in the north (Yao et al., 2016). It is the most severe pathogen in the agricultural sector and causes blackening of the vegetable roots with subsequent wilting and necrosis in the earlier stage and plant death in the later stages, resulting in a >20% loss in harvest yield.
Continuous cropping is common in agricultural production and can lead to a series of issues such as degradation of soil quality, aggravation of diseases and pests, and reductions in crop yield (He et al., 2008; Wu et al., 2015). It is the main reason for the spread of soil-borne diseases due to nutrient imbalances, decreases in enzyme activity, and physical/chemical and microflora disturbances (Li et al., 2016). Soil microbes play key roles in soil micro-ecology such as organic matter decomposition, nutrient transport, purification of environmental pollutants, and mediation of greenhouse gases (Kennedy and Smith, 1995; Berendsen et al., 2012; Zhao et al., 2015). Soil microbial activity and respiration intensity are closely related to soil catalase activity, which is an important indicator of the soil micro-ecological environment. Li et al. (2016) reported that the relative abundance of dominant bacteria increases considerably with long-term consecutive monoculture in the rhizosphere of black pepper, while an increasing trend has been observed in the abundance of pathogenic Fusarium fungi paired with a decrease in Pseudomonas and Bacillus bacteria. Similarly, long-term continuous cropping causes a significant decrease in soil pH and organic matter enzymatic activities and results in bacterial decline in black pepper soil (Xiong et al., 2015). This promotes soil microbial imbalances and reduces the number of beneficial microbes while increasing pathogenic microorganisms, leading to a higher susceptibility of plants to various soil-borne diseases. The plant’s ability to counter soil-borne disease and the activity of soil enzymes are closely related to the composition of microbes present in the rhizosphere (Magnuson and Crawford, 1992; Simoes et al., 1997). Therefore, improving the soil environment is critical for countering the detrimental effects of continuous cropping. However, there are few studies on the effect of continuous pepper cropping on the microbial community composition in the rhizosphere over time.
The main methods of prevention and control of pepper blight are the use of resistant pepper varieties and chemical agents. However, the lack of resistant varieties and the problems of using chemical agents limit the prevention and control of pepper blight(Hartman and Wang, 1992; Hai et al., 2013; Barchenger et al., 2018). Therefore, biological control has become an attractive option and has been studied by many researchers, in which screening for bacteria that have the ability to control pepper blight is a key process (Sid et al., 2003; Wang et al., 2019). Many microorganisms with the ability to control pepper blight have been isolated from plants and rhizosphere soil including Bacillus sp strains, Trichoderma harzianum strains, and Actinomycetes sp strains (Anitha et al., 2003; Tran et al., 2008). Twelve isolates have been obtained from the pepper rhizosphere, and one strain, named PS119, has a remarkable ability to suppress the growth of P. capsici (Rajkumar et al., 2005). Aspergillus sp strains are widely distributed saprophytic fungi that have been used industrially in processes such as alcohol fermentation, biological engineering and transformation, and genetic research to produce antibiotics, organic acids, and enzymes (Berka et al., 1992; Roehr et al., 1992; Aybeke et al., 2014; Park et al., 2017). However, there are limited reports of using Aspergillus sp strains to control pepper blight.
To determine the microbial community and diversity in continuously cropped pepper soil, we selected soil that had been subjected to continuous cropping for 3, 6, and 9 years and identified the number of bacteria, fungi, and actinomycetes in the rhizosphere by dilution plating. The morphological, physiological, and biochemical characteristics as well as 16S ribosomal DNA (rDNA) or rDNA internal transcribed spacer (ITS) sequences were examined to study the soil microbial ecology. These findings provide the foundation for optimising the soil micro-environment to control pepper blight.
Materials and Methods
Rhizosphere Soil Collection
The experimental soil samples were collected from different long-term pepper greenhouse fields that were consecutively cropped for 3, 6, and 9 years with the same agronomic management and fertilisation regime in Beizhen city, Liaoning province of China. Soil that was less than 5 mm from the root surface was gathered and defined as the rhizosphere. The rhizosphere soil of healthy and diseased pepper plants was collected in the same greenhouse, and six treatments were tested. Three greenhouses were randomly selected for each pepper planting year. The five-point sampling method was used in each greenhouse (Xin et al., 2006), and for each pepper field, five healthy and P. capsici-infected pepper plants were randomly collected with intact roots and surrounding soil by shaking (Dong et al., 2013). Each treatment with three replicates included 15 sample sites and 75 rhizosphere soil of pepper. All soil samples were rapidly frozen with liquid nitrogen and stored at -80°C for DNA extraction.
Culture Media
Luria-Bertani (LB) media was used for bacteria isolation and culture, Potato-Martin substratum was used for fungi isolation and culture, and Gao No. I media was used for actinomycetes isolation and culture. The compositions of the media are shown in Suppl. Table 1s (Fang, 1998; Dong and Cai, 2001).
Soil Microbial Isolation and Purification
Dilution plating was used to isolate bacteria, fungi, and actinomycetes from different long-term consecutively cropped soils. The ratio of microbes between healthy and diseased soil was calculated as follows: R (%) = (N1-N2)/N2 × 100% where N1 represents the amount of bacteria, fungi, and actinomycetes in diseased soil and N2 is the amount in healthy soil.
Physiological and Biochemical Identification
The physiological and biochemical index, catalase reaction, methyl red (MR) reaction, V-P reaction, starch hydrolysis reaction, cellulose decomposition, nitrate reduction, nitrite reduction, H2S, lipase, tryptophan deaminase enzyme, phenylalanine deaminase, citrate utilization, and salt tolerance were examined for physiological and biochemical identification of bacteria and actinomycetes.
DNA Extraction and Illumina Sequencing
The DNA extraction kits for bacteria and fungi (Tiangen Biotech, China) were used to extract bacterial and fungal DNA, which was examined by 1.2% agarose gel electrophoresis. The OD260 and OD280 values were also measured, and OD260/280 was calculated to determine the concentration and purity of DNA. We used 2× Taq PCR Master Mix kits (Tiangen Biotech, China) for PCR amplification, after which we sequenced the amplification products. Each reaction contained 12.5 µL master mix, 1.0 µL DNA, 0.5 µL of each primer, and 14 µL sterile distilled water. The PCR cycle was as follows: 94°C denaturation for 3 min, 94°C denaturation for 30 sec, 55°C annealing for 30 sec, and 72°C extension for 1 min for 30 cycles followed by 72°C extension for 5 min. Bacteria and actinomycetes 16S rDNA PCR primers were 5'-AGAGTTTGATCCTGGCTCAG-3' (forward) and 5'-GGTTACCTTGTTACGACTT-3' (reverse). Fungal rDNA-ITS PCR primers were 5'-TCCTCCGCTTATTGATATGC-3' (forward) and 5'-GGAAGTAAAAGTCGTAACAAGG-3' (reverse).
Sequence Alignment and Cluster Analysis
The obtained sequences were blasted in the NCBI database. Clustalx1.83 was used to construct an alignment, and MEGA 4.0 was used to infer phylogenetic trees. Bacteria and actinomycetes were identified by colony and cell morphology, gram staining, physiological and biochemical tests, and 16S DNA sequence analyses. Fungi were identified by colony and cell morphology and rDNA-ITS sequence analysis.
Results
Soil Micro-Ecology
Bacteria including actinomycetes and fungi in the soil drive the soil ecosystem’s biochemical cycle (Cao et al., 2011). To understand the effects of continuous cropping of pepper on the rhizosphere soil ecosystem over time, we studied the number of cultivatable bacteria, actinomycetes, and fungi in farms after 3, 6, and 9 years (3a, 6a, and 9a, respectively) of continuous cropping (Table 1). The number of bacteria in the rhizosphere of healthy plants at 3a, 6a, and 9a was significantly higher than in diseased soil and showed a peak at 6a. We did not observe a significant difference in bacteria quantity in healthy plants at 3a, 6a, and 9a or diseased plants at 3a and 9a. However, the number of bacteria in diseased plants at 6a was significantly elevated. In addition, a comparison between the number of rhizosphere bacteria in healthy and diseased plants in the same year showed consistently lower bacteria counts at all time points. We observed the largest decrease in diseased plants at 9a followed by the 3a and 6a time points. Similarly, the number of rhizosphere actinomycetes in healthy plants at 3a, 6a, and 9a was slightly higher than in diseased soil but showed an inverse temporal trend compared with bacteria. Furthermore, the number of rhizosphere actinomycetes was significantly increased at 9a in healthy plants, while we did not detect any significant differences in diseased plants. Actinomycete counts at 3a, 6a, and 9a in diseased plants were 7.67%, 5.42%, and 12.35% lower, respectively, compared with healthy plants. Compared with rhizosphere bacteria, the decrease in actinomycetes in diseased plants was relatively mild. Conversely, the number of fungi in the rhizosphere of diseased plants was significantly higher than in healthy plants with a peak at 6a. We observed increases of 37.82%, 19.75%, and 17.29% at 3a, 6a, and 9a, respectively. In healthy plants, the number of fungi was significantly elevated at 6a compared with 3a, but no difference was found between 6a and 9a. However, in diseased plants, the fungi count continued to rise significantly at 9a.
Table 1.
Quantity of soil microbes in the continuously cropped soil of healthy and diseased pepper plants
| Consecutive years |
Average number of Bacteria (108 CFU/mL) |
Average number of Actinomycetes (106 CFU/mL) |
Average number of Fungi (104 CFU/mL) | |||||
| Healthy plants | Diseased plants | Healthy plants | Diseased plants | Healthy plants | Diseased plants | |||
| 3 a | 3.44 ± 0.36 c | 0.64 ± 0.13 b | 3.13 ± 0.33 b | 2.89 ± 0.27 a | 8.09 ± 0.37 b | 11.15 ± 0.45 c | ||
| R (%) | 537.50 | 108.31 | 72.56 | |||||
| 6 a | 5.55 ± 0.31 a | 1.19 ± 0.16 a | 2.95 ± 0.46 b | 2.79 ± 0.23 a | 12.16 ± 0.50 a | 19. 75 ± 0.82 a | ||
| R (%) | 466.39 | 105.74 | 61.57 | |||||
| 9 a | 4.49 ± 0.33 b | 0.48 ± 0.10 b | 4.29 ± 0.33 a | 3.76 ± 1.25 a | 11.89 ± 0.40 a | 17.29 ± 0.42 b | ||
| R (%) | 935.42 | 114.10 | 68.77 | |||||
The bacteria/fungi ratio reflects the structure and nutritional function of the soil food chain. We found significantly higher bacteria/fungi ratio at 3a, 6a, and 9a in healthy plants compared with diseased plants (Table 2), which decreased by 761.67%, 766.67%, and 1266.67%, respectively. The decrease was highest at 9a, indicating that the growth rate of bacteria was significantly higher than that of fungi. In both healthy and diseased soil, bacteria dominated in absolute numbers followed by actinomycetes and fungi (Tables 1 and 2). Taken together, the number of bacteria and actinomycetes in the rhizosphere of healthy plants was higher than in diseased plants, while the number of fungi was lower. Infection by P. capsici had the greatest impact on the number of soil bacteria and the least impact on fungi and actinomycetes. Time point 6a appeared to be the pivotal point at which the microbial composition changed significantly.
Table 2.
Proportion of bacteria, fungi, and actinomycetes in the continuously cropped soil of healthy and diseased pepper plants
| Consecutive years | B/F (104) | A/F (102) | B/A (102) | |||||
| Healthy plants | Diseased plants | Healthy plants | Diseased plants | Healthy plants | Diseased plants | |||
| 3 a | 0.43 ± 0.05 a | 0.06 ± 0.01 a | 0.39 ± 0.06 b | 0.26 ± 0.03 a | 0.92 ± 0.15 a | 4.52 ± 0.53 b | ||
| R (%) | 716.67 | 150.00 | 20.36 | |||||
| 6 a | 0.46 ± 0.02 a | 0.06 ± 0.01 a | 0.24 ± 0.05 a | 0.14 ± 0.01 b | 0.53 ± 0.10 b | 2.34 ± 0.47 c | ||
| R (%) | 766.67 | 171.43 | 22.65 | |||||
| 9 a | 0.38 ± 0.04 a | 0.03 ± 0.01 b | 0.36 ± 0.02 b | 0.22 ± 0.07 ab | 0.96 ± 0.16 a | 7.90 ± 1.33 a | ||
| R (%) | 1266.67 | 163.64 | 12.15 | |||||
Colony Characteristics and Microscopic Morphology of Bacteria, Actinomycetes, and Fungi in the Soil of Continuously Cropped Pepper
Bacteria were isolated from soil by dilution plating, and the morphology of the main colony is shown in Table 3. We performed a Gram stain and observed the microscopic morphology using a microscope at ×100 magnification (Table 4).
The isolated actinomycetes with dry, wrinkled, and concentric growth characteristics were selected, inoculated on actinomycete culture medium, and cultured at 28°C for 5–7 d. The colony morphology was then noted (Table 3). Glass slides were inserted into the actinomycete culture medium to examine the spore morphology after 7 d (Table 4). Fungi were isolated from soil, inoculated on PDA medium, and incubated at 25°C for 5–7 d. The colony morphology was then observed (Table 3), and the mycelia were isolated onto plates and examined using an optical microscope (Table 4).
Table 3.
Morphology of bacterial, actinomycete, and fungal colonies in the soil of continuously cropped pepper
Table 4.
Microscopic morphology of bacteria, actinomycetes, and fungi in the soil of
continuously cropped pepper
Physiological and Biochemical Properties of Bacteria and Actinomycetes in the Soil of Continuously Cropped Pepper
A set of 15 bacteria and 17 actinomycetes were selected for physiological and biochemical identification and examined by physiological and biochemical indexes, hydrogen peroxide production, MR, V-P, starch and cellulose hydrolysate content, and gelatine liquefaction ability (Table 5).
Table 5.
Physiological and biochemical properties of bacteria and actinomycetes in the soil of continuously cropped pepper
Sequencing and Cluster Analysis of Bacteria, Actinomycetes, and Fungi in the Soil of Continuously Cropped Pepper
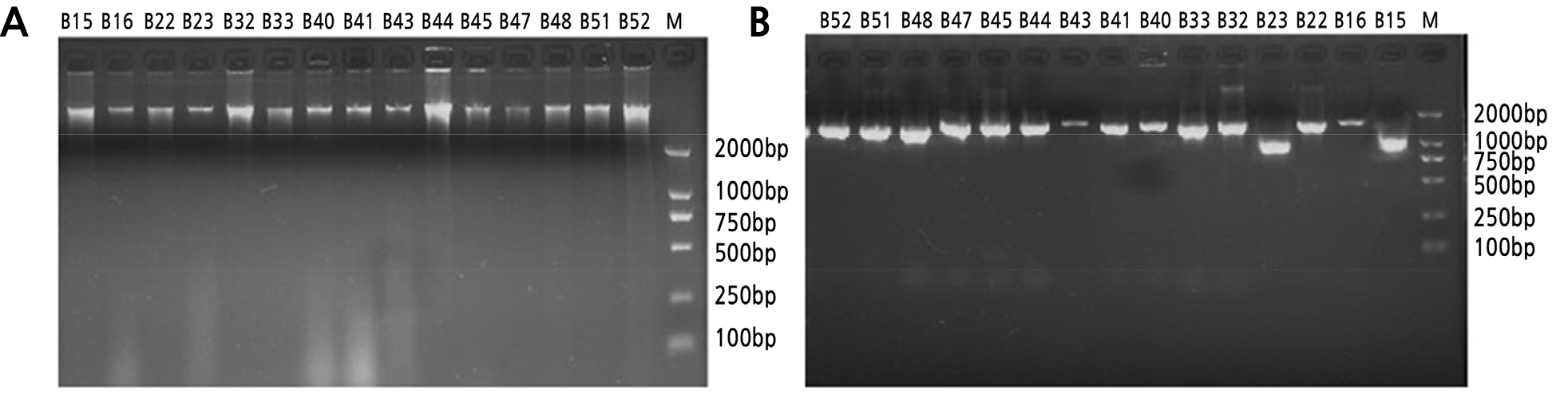
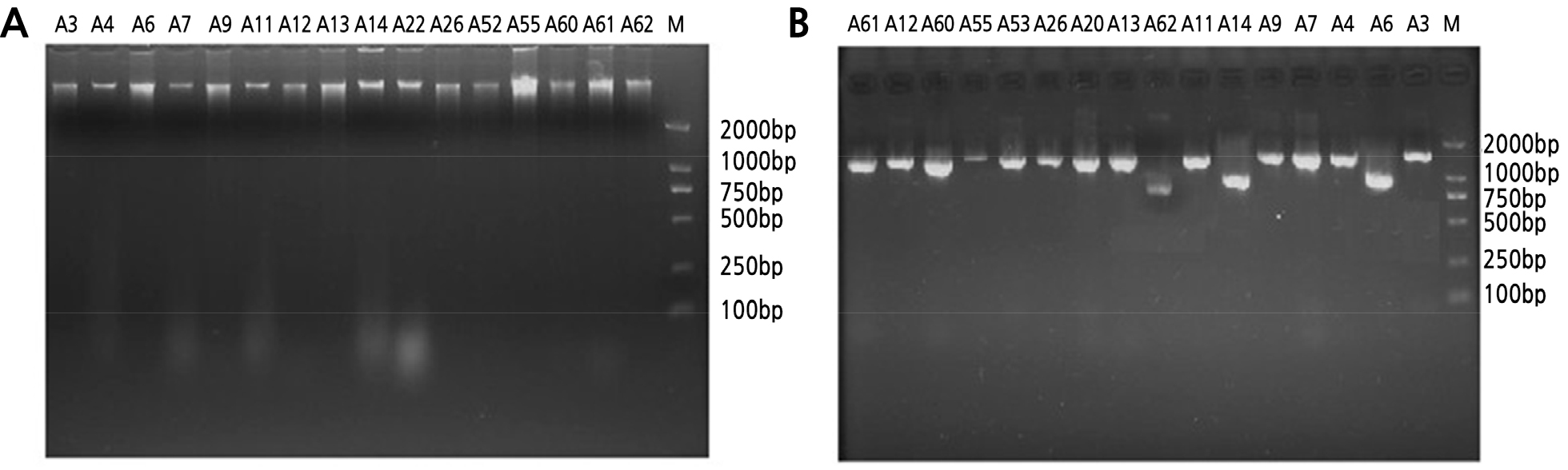
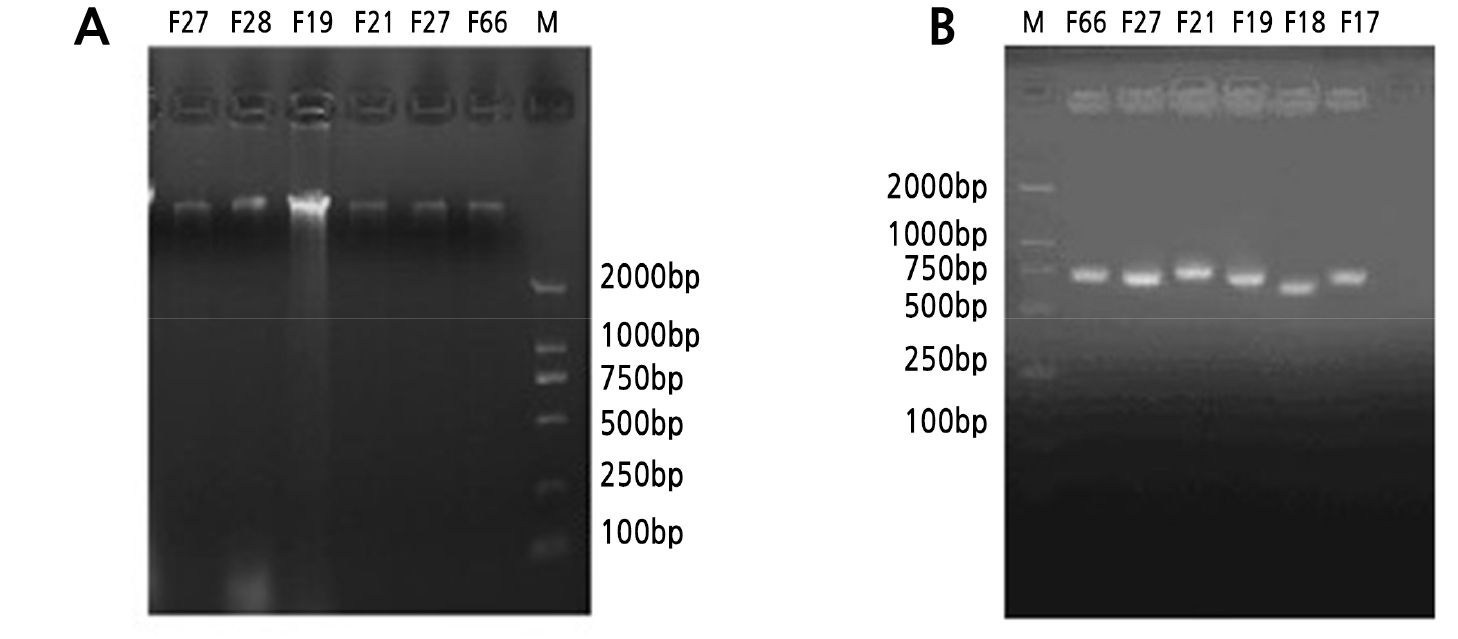
Identification was performed according to the common bacteria system identification manual. Bacterial, actinomycete, and fungal DNA was extracted and detected by 1.2% agarose gel electrophoresis (Figs. 1A, 2A and 3A). We calculated an OD260/280 value of 1.8 to 2.0 and performed 16S rDNA PCR. The results indicated that the size of the bacterial DNA amplification sequences B22 and B15 were about 1000 bp, and the remaining sequences were 1500 bp (Fig. 1B). In comparison, actinomycete and fungi sequences were 2000 bp long (Figs. 2B and 3B). We then sequenced the PCR products and found that the nucleotide fragments of bacteria, actinomycetes, and fungi were 952–1466 bp, 948–1440 bp, and 500–750 bp, respectively (Table 6). The sequences were compared using the NCBI database, and Clustalx1.83 was used to construct an alignment. MEGA 4.0 was employed for cluster analyses of the respective bacteria and actinomycetes.
Table 6.
Sequencing and cluster analysis of bacteria, actinomycetes, and fungi in the soil of continuously cropped pepper
Discussion
Bacteria play an important role in changing soil fertility and structure and promoting nutrient circulation. In this study, we showed that the number of bacteria and actinomycetes over time in the rhizosphere of healthy and P. capsici-infected pepper plants first increased followed by a decrease. Other studies have observed that the bacterial communities in the rhizosphere of Rehmannia glutinosa significantly decrease in quantity and diversity with continuous cropping, which impairs the growth of R. glutinosa and its underground tubers and reduces its usability as a medicinal ingredient (Zhang et al., 2010). In addition, Wu and Wang (2007) and Qin et al. (2015) observed similar trends in soil bacteria from continuously cropped cucumber and potato (Wu and Wang, 2007; Qin et al., 2015). However, the results of our research showed some discrepancies with these studies, possibly due to the prolonged period of continuous cropping of up to 9 years included in our study. Vegetable farmers increase the application of fertilisers and pesticides at the early stages of discovering P. capsici infection, which artificially changes the original nutrients present in the soil followed by adaptations in the number of soil microorganisms and the structure of the flora (Yang et al., 2000; Van Schoor et al., 2009; Meng et al., 2012; He et al., 2014). We showed that after infection, the pepper plant’s rhizosphere bacteria decreased significantly, whereas actinomycetes showed no significant changes and the number of fungi increased. These observed changes in bacteria and fungi were consistent with barley root rot (Li et al., 2017), tobacco bacterial wilt (Li et al., 2020), and banana wilt (Deng et al., 2011). An explanation for this phenomenon is that the number of pathogenic fungi in the soil suddenly increases after disease and directly competes with the resident bacteria, actinomycetes, and fungi in the soil for nutrition and space, which leads to a decline in the number of beneficial microorganisms and changes the soil from “bacterial type” to “fungal type.”
The stark changes in the ratio of bacteria/fungi and actinomycetes/fungi in this study all appeared in the sixth year. This is possibly due to shifts in the absorption ability and viability of the pepper roots after prolonged continuous cropping, leading to different compositions in root exudates. This may contribute to altering the composition of microorganisms in the soil and the subsequent detrimental effects. Similar observations have been reported previously. Zhu et al. (2019) studied the number of microorganisms in the rhizosphere soil of tobacco after 2, 4, and 6 years of continuous cropping and found that 4 years is the pivotal point when the number of bacteria, actinomycetes, and fungi in the soil change significantly (Zhu et al., 2019). Furthermore, Wu and Wang (2007) reported that the soil’s microbial abundance and diversity of continuously cropped cucumber after 7 years is significantly lower than after 2 years (Wu and Wang, 2007). Crop rotation is a means of improving the soil’s micro-ecological environment and increases the yield and quality of agricultural products. Dong et al. (2019) found that when garlic and maize are rotated with pepper, the soil’s microbial community structure and diversity is improved paired with a reduction in the threat of pathogens (Dong et al., 2019). Zhang et al. (2015) found that cucumber and leafy vegetable rotation increases the number of soil microorganisms and reduces soil salinity (Zhang et al., 2015). Therefore, the data of our study suggest that when peppers are continuously planted for 5–6 years, it would be beneficial to rotate them with different crops such as garlic or leafy vegetables (Dong et al., 2019), thus improving the soil’s nutrients and increasing enzyme activity and microorganism diversity. These factors would ultimately promote the healthy and sustainable agriculture of peppers.
Seven of the 15 strains of bacteria obtained in this study belonged to the genus Bacillus thuringiensis. This is likely because spores are dormant bodies of bacteria, which are among the most stress-resistant and adaptable organisms that can survive adverse external environments such as extreme temperatures, absence of water or light, and presence of chemicals (Bağcıoğlu et al., 2019). Therefore, Bacillus can exist in large quantities when the soil’s micro-ecological environment is destroyed by continuous cropping. Bacillus also has many ecological functions such as enzyme production, salt tolerance, acid production, and phosphorus solubilisation (Céline et al., 2007). For example, Paenibacillus polymyxa secretes polypeptide proteins, enzymes, and plant hormones, which may be used to control plant diseases (Lin et al., 2018) and can promote plant growth by fixing nitrogen and dissolving phosphorus (Khan et al., 2008). Others, such as Bacillus amyloliquefaciens, secrete proteins, lipopeptides, and secondary metabolites to inhibit the growth of plant pathogenic bacteria (Yan et al., 2018). Streptomyces is the most diverse genus in the phylum of actinomycetes (Dai et al., 2014). It can adapt to different environments through a variety of self-produced secondary metabolites (Zeng et al., 2019; Ma et al., 2019). Therefore, of the 17 actinomycete species isolated in this study, 15 belonged to the genus Streptomyces. Additionally, Streptomyces can produce a variety of secondary metabolites such as aminoglycosides, nucleosides, polyenes, macrolides, hormones, and tetracyclines, which are important for cell wall and protein synthesis, plants growth, and the creation of an acidic environment that inhibits pathogens (Barakate et al., 2002; Wang et al., 2005; Hamedi and Mohammdipanah, 2015). Therefore, Streptomyces may be valuable for biological control of plant disease and promotion of plant growth. Among the six isolated fungi, Phytophthora, Fusarium solani, and Fusarium were the pathogenic microorganisms for Phytophthora capsica infection, root rot, and fusarium wilt, respectively, indicating that continuous cropping led to the domination of pathogenic fungi over beneficial bacteria. As a saprophytic fungus, aflatoxin produced by Aspergillus is widely distributed in the soil, which does not have a high requirement for growth and is an opportunistic plant pathogen (Horn, 2003). Penicillium oxalate promotes disease resistance by inducing plant resistance and phosphorus solubilisation (Fan et al., 2002; Peng et al., 2004; Ali et al., 2006). Penicillium expansum, on the other hand, can cause postharvest rot of apples and pears and even human intestinal diseases (Arici et al., 2000; Liu et al., 2010). We found that after continuous cropping, the majority of the bacteria other than Penicillium were pathogenic. The dominant bacterial strains isolated in this study, such as Bacillus, Streptomyces, and Penicillium, are beneficial and available microbial resources.
This research was executed using traditional isolation and culture methods. Compared with modern high-throughput molecular sequencing technologies, traditional methods lack versatility and comprehensiveness and can only be applied on cultivatable microorganisms in the soil, thus limiting the scope of the research (Yuan et al., 2014). However, modern molecular biology technology does not aim to obtain living microbial cells that can be cultured, making it difficult to accurately design and efficiently utilise these microorganisms to fine-tune microbial processes in the soil. In this aspect, the traditional pure culture technology can isolate and obtain pure microorganisms that may be directly or indirectly used for research, medicine, industry, and agricultural production through expansion and cultivation or transformation into new strains (Guo et al., 2006). In conclusion, our study provides a basis for human intervention to beneficially alter the soil microbial flora after continuous cropping to break the current limitations and sustainably guide agricultural production.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20210039_Table_1s.pdf
The composition of medium