Introduction
Materials and Methods
Plant Materials
Microscopic Observations of the Mixed Floral Buds
Acquirement of Gene Sequence
Phylogenetic and Motif Analyses
Quantitative Real-Time PCR
Results
Identification and Classification of MADS-box Genes in Chinese Chestnut
Phylogenetic Analysis of MADS-box Proteins
Conserved Motifs of MADS-box Proteins
Expression Analysis of CmMADS Genes in Reproductive Organs
Discussion
Introduction
MADS proteins play a crucial role in many important processes in flowering plants, such as floral bud initiation, flowering time control, floral organ identity, fruit development, and the vegetative to reproductive phase transition (Liu et al., 2012; Lu et al., 2012; Goldberg-Moeller et al., 2013). MADS proteins are characterized by the presence of a highly conserved MADS-box domain that has approximately 60 amino acids located in the N-terminal region (Shore and Sharrocks, 1995; Riechmann et al., 1996; Kaufmann et al., 2005; Shu et al., 2013). The MADS-box genes were divided into two subfamilies, type I and type II MADS genes, based on the conserved domain of MADS proteins (Xu et al., 2014). Type II genes, also known as the MIKC type, encode proteins that contain the MEF2-like MADS, I, K, and C domains. The MEF2-like MADS domain is located at the N-terminus region and has the most conserved structure (De et al., 2003; Immink et al., 2002). The I and K domains, two intervening regions, are located downstream of the MEF2-like MADS domain, but their sequence and length vary significantly (Parenicová et al., 2003). In contrast, the C domain shows low conservation with high diversity (Honma and Goto, 2001). Compared with type II genes, type I genes are a primitive type, often containing one or two exons and lack a K domain (Alvarez-Buylla et al., 2000; Smaczniak et al., 2012).
Type I proteins are divided into Mα, Mβ, and Mγ groups, and type II proteins are divided into groups MIKCC and MIKC* (Henschel et al., 2002). In addition, MIKCC genes are divided into 13 subgroups, including the AP1, AP3, AG, SEP, FLC, SOC1, SVP, AGL6, AGL15, AGL17, AGL12, BS, and TM8 subgroups (Becker and Theissen, 2003). The ‘floral quartet model’, involving the ‘ABCDE’ genes, describes how specific genes from the AP1, AP3, AG, and SEP subgroups are responsible for the formation of sepals, petals, stamens, carpels, and ovules in Arabidopsis thaliana (Kaufmann et al., 2005; Wells et al., 2015). In turn, the FLC, SOC1, and SVP subgroup genes regulate the vernalization and flowering time in some plant species (Amasino, 2005; Hemming et al., 2011). Type I genes are involved in female gametophyte and seed development in A. thaliana (Bemer et al., 2010). Analysis of the MADS gene family in woody plants, such as poplar (Populus trichocarpa) (Leseberg et al., 2006), sweet orange (Citrus sinensis) (Hou et al., 2014), mei (Prunus mume)(Xu et al., 2014), peach (Prunus persica) (Wells et al., 2015), apple (Malus domestica) (Tian et al., 2015), grapevine (Vitis vinifera) (Grimplet et al., 2016), and Chinese jujube (Ziziphus jujube) (Zhang et al., 2017), have offered a good research basis into the MADS genes in woody plants.
Chinese chestnut (Castanea mollissima) is a woody plant species with important economic and ecological value and is widely cultivated for nut production (Feng et al., 2011; Guo et al., 2012). As a monoecious plant, Chinese chestnut exhibits two types of catkins: a unisexual catkin with a purely staminate cluster and a bisexual catkin that has both staminate and pistillate flowers (Roberto et al., 1995; Mert and Soylu, 2006; Feng et al., 2011; Warmund, 2011). The ratio of female to male flowers is approximately 1:2900-3400, and the production of fewer female flowers significantly limits chestnut yield (Guo et al., 2012). Therefore, the identification of genes related to female flower development is extremely important in Chinese chestnut, and MADS-box genes are directly involved in floral organ morphogenesis. Therefore, here, we studied the roles that MADS-box genes play in floral bud initiation and floral organ morphogenesis in Chinese chestnut. Previous studies have identified 19 differentially expressed MADS-box genes from female and male flowers of the Chinese chestnut mixed inflorescence (Liu et al., 2006; Qiu et al., 2016; Gao et al., 2020). In this study, we identified MADS-box proteins in Chinese chestnut and examined their phylogeny, motif composition, and gene expression. The results provide valuable insight for further functional studies of CmMADS genes and lay a theoretical foundation for understanding the regulation of Chinese chestnut flower development, especially with regard to promoting female flower development, and contribute to genetic breeding improvement in Chinese chestnut.
Materials and Methods
Plant Materials
Samples were collected from 15-year-old individuals of the Chinese chestnut (Castanea mollissima) cultivar ‘Yanshanhongli’ at the Chestnut Experiment Station in Beijing, China. Mixed floral buds were harvested during the inflorescence primordium early differentiation stage (March 14, 2016); mixed floral bud development stages B1-B4 were harvested during the inflorescence primordium early differentiation stage (March 14, 2016), the inflorescence primordium later differentiation stage (March 25, 2016), the floral clusters bract primordium formation stage (April 8, 2016), and the floral clusters primordia shaped formation stage (April 22, 2016). Morphological observations of floral organs are shown in Fig. 1. The leaf and cotyledon were normal and mature. All samples were collected for three biological replicates, immediately frozen in liquid nitrogen, and stored at -80°C. The mixed floral bud development stages B1-B4 were fixed for paraffin sectioning by formalin/acetic acid/70% ethanol (FAA) at a ratio of 5:5:90.
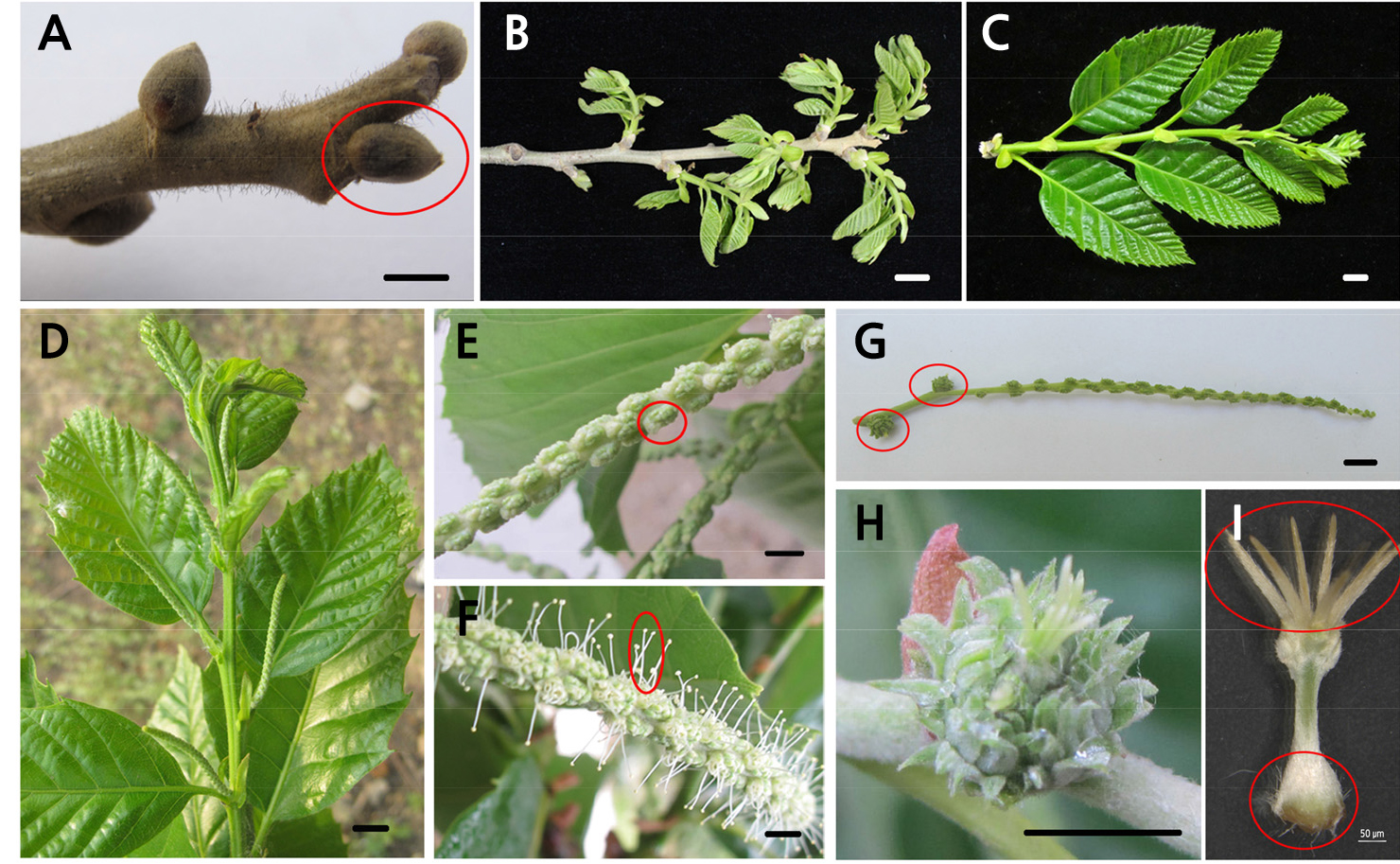
Fig. 1.
Morphological observation of floral organs. (A) Growth and development of mixed floral buds of fruiting branches; the red circle represents the mixed floral bud, (B) Development of the fruiting branches, (C) Development of annual branch, (D) Growth and development of staminate catkin, (E) Male floral cluster in catkin, (F) Growth and development of stamen in male floral cluster, (G) Growth and development of early female flower cluster; the red circles represent two female flowers from a bisexual catkin, (H) Growth and development of late female flower cluster, (I) Stigma and ovary in the female flower; the upper red circle represents the stigma, and the lower red circle represent the ovary. Scale bars: A-H = 1 cm; I = 50 µm.
Microscopic Observations of the Mixed Floral Buds
The mixed floral buds were microscopically observed using the paraffin sections by the protocol in Wang et al. (2019). The mixed floral buds were dehydrated using an ethanol series (50-100%) and xylene (50 and 100%). Then, the samples were infiltrated with paraffin (24 h, 60°C) and cut into 10-µm-thick sections. The sections were stained with safranin solution and observed by an Olympus BX-51 optical microscope.
Acquirement of Gene Sequence
The Chinese chestnut gene and protein sequences were obtained from Chinese chestnut genome project (http://gigadb.org/ dataset/100643) (Xing et al., 2019). Candidate CmMADSproteins containing the SFR (PF00319) and the MEF2 (PF09047) domains were searched using BLASTP searches against the genome sequence in Chinese chestnut. Sequences lacking MADS domains were removed in subsequent analyses using the Pfam (http://Pfam.sanger.ac.uk/) and National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) databases. The characterization parameters were calculated using the ExPASyz online tools (http://web.expasy.org/protparam/).
Phylogenetic and Motif Analyses
Phylogenetic dendrograms were generated using the MEGA 5.01 tool, and the MADS protein sequences (listed in the supplemental information) were aligned by Clustal X2.0 with default parameters. The neighbor-joining trees using the MADS-box proteins were constructed and bootstrap values were calculated for 1000 iterations (Larkin et al., 2007; Tamura et al., 2011). The phylogenetic relationship of the TM8 gene was constructed for multiple species and all gene and protein sequences for the phylogenetic trees are listed in the supplemental information. The motif analysis of CmMADS proteins was performed using the MEME online tool (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2009). The following parameters were used: number of repetitions, any; maximum number of motifs, 10; minimum motif width, 6; and maximum motif width, 60.
Quantitative Real-Time PCR
Total RNA was extracted separately from Chinese chestnut samples using a Plant RNA Kit (Omega, USA). cDNA was synthesized using M-MLV reverse transcriptase (Takara, Japan). The protocols used for quantitative real-time PCR (qRT-PCR) and primer design were based on the study of Chen et al. (2019). The qRT-PCR analysis was performed by a LightCycler 96 Real-Time PCR with SYBR Green (Takara, Japan). The primers for the CmMADS genes are listed in Suppl. Table 1s. Each qRT-PCR was carried out in a volume of 10 µL, which contained 5 µL of SYBR Premix Ex Taq (2×), 2.5 µL of ddH2O, 2 µL of diluted template (60-fold dilution of cDNA), and 0.25 µL of each gene-specific primer. The following reaction program was used: 95°C for 10 min, followed by 39 cycles at 95°C for 20 s, the annealing temperature based on each pair of primers for 20 s, and 72°C for 20 s. All qRT-PCR experiments were performed with three biological replicates. To verify the specificity of each primer, melting curve analysis was performed from 60°C to 95°C. The raw cycle threshold (Ct) values were calculated using the 2-ΔCt method, and CmActin was employed as a reference gene, which maintained almost constant expression levels under all experimental conditions. The mean relative expression value of CmMADS genes in floral organs are listed in Suppl. Table 2s and Suppl. Table 3s. The heat maps were generated based on the mean relative expression value using the HemI 1.0.3.3 tool.
Results
Identification and Classification of MADS-box Genes in Chinese Chestnut
In total, 53 MADS-box genes were identified in the Chinese chestnut genome; 21 of which were classified as type I MADS-box genes and 32 as type II MADS-box genes. The characterization parameters of CmMADS proteins are shown in Table 1, with a deduced length ranging from 98 (CmMADS01) to 540 amino acids (CmMADS45), a predicted molecular mass ranging from 11281.86 (CmMADS01) to 61,665.78 (CmMADS45) Daltons (Da), and isoelectric point (pI) values ranging from 5.20 (CmMADS42) to 10.39 (CmMADS01).
Table 1.
The MADS-box proteins identified in Chinese chestnut
| Proposed name | Type | Len | MW (Da) | pI | Ai | GRAVY | Instability index (II) |
| CmMADS01 | Mα | 98 | 11281.86 | 10.39 | 42.86 | -0.915 | 74.18 |
| CmMADS02 | Mα | 164 | 19062.57 | 6.24 | 60.67 | -0.790 | 61.04 |
| CmMADS03 | Mα | 150 | 17183.13 | 9.68 | 78.60 | -0.495 | 52.14 |
| CmMADS04 | Mα | 217 | 25276.08 | 7.62 | 65.07 | -0.661 | 65.73 |
| CmMADS05 | Mα | 217 | 25295.04 | 6.85 | 64.61 | -0.682 | 61.47 |
| CmMADS06 | Mα | 250 | 28300.87 | 6.26 | 33.45 | -0.817 | 33.45 |
| CmMADS07 | Mα | 220 | 24791.28 | 8.40 | 68.64 | -0.647 | 34.18 |
| CmMADS08 | Mα | 187 | 20864.06 | 9.76 | 77.70 | -0.617 | 40.55 |
| CmMADS09 | Mα | 214 | 24095.36 | 8.94 | 61.96 | -0.743 | 50.10 |
| CmMADS10 | Mα | 217 | 24108.21 | 8.76 | 63.41 | -0.790 | 52.25 |
| CmMADS11 | Mα | 199 | 21543.15 | 5.98 | 74.67 | -0.508 | 36.41 |
| CmMADS12 | Mα | 226 | 25385.03 | 6.84 | 88.50 | -0.420 | 42.79 |
| CmMADS13 | Mα | 220 | 24654.18 | 9.68 | 75.82 | -0.580 | 36.49 |
| CmMADS14 | Mα | 209 | 23540.89 | 8.30 | 90.10 | -0.391 | 36.43 |
| CmMADS15 | Mα | 219 | 24761.37 | 9.63 | 84.66 | -0.437 | 42.70 |
| CmMADS16 | Mβ | 337 | 38964.74 | 8.69 | 70.27 | -0.604 | 54.73 |
| CmMADS17 | Mβ | 327 | 38176.94 | 9.18 | 67.95 | -0.673 | 47.43 |
| CmMADS18 | Mβ | 433 | 49017.78 | 8.84 | 74.48 | -0.520 | 37.36 |
| CmMADS19 | Mγ | 406 | 43669.18 | 7.75 | 54.26 | -0.628 | 38.30 |
| CmMADS20 | Mγ | 253 | 29055.41 | 8.97 | 71.30 | -0.698 | 52.95 |
| CmMADS21 | Mγ | 256 | 29409.71 | 9.30 | 60.59 | -0.779 | 43.34 |
| CmMADS22 | MIKCC | 171 | 19700.63 | 9.37 | 93.04 | -0.674 | 41.84 |
| CmMADS23 | MIKCC | 240 | 27562.33 | 8.86 | 83.29 | -0.681 | 48.27 |
| CmMADS24 | MIKCC | 295 | 33160.46 | 9.47 | 83.63 | -0.630 | 47.93 |
| CmMADS25 | MIKCC | 244 | 27930.66 | 9.10 | 82.34 | -0.679 | 40.47 |
| CmMADS26 | MIKCC | 266 | 30816.07 | 9.15 | 78.46 | -0.890 | 50.79 |
| CmMADS27 | MIKCC | 248 | 28275.23 | 9.01 | 80.24 | -0.635 | 54.89 |
| CmMADS28 | MIKCC | 242 | 27920.71 | 9.59 | 78.60 | -0.777 | 52.65 |
| CmMADS29 | MIKCC | 186 | 21444.75 | 10.23 | 86.45 | -0.620 | 41.10 |
| CmMADS30 | MIKCC | 200 | 22965.31 | 9.43 | 87.80 | -0.614 | 51.80 |
| CmMADS31 | MIKCC | 222 | 25466.25 | 9.51 | 89.59 | -0.554 | 51.82 |
| CmMADS32 | MIKCC | 170 | 19707.49 | 9.60 | 78.06 | -0.814 | 55.05 |
| CmMADS33 | MIKCC | 246 | 28511.42 | 8.71 | 80.53 | -0.767 | 56.85 |
| CmMADS34 | MIKCC | 243 | 27795.87 | 9.03 | 83.05 | -0.617 | 62.90 |
| CmMADS35 | MIKCC | 207 | 23935.36 | 7.63 | 84.78 | -0.669 | 64.68 |
| CmMADS36 | MIKCC | 202 | 23361.93 | 9.61 | 82.52 | -0.856 | 57.51 |
| CmMADS37 | MIKCC | 200 | 22936.68 | 6.85 | 94.15 | -0.377 | 57.31 |
| CmMADS38 | MIKCC | 204 | 23426.77 | 10.00 | 86.47 | -0.710 | 66.25 |
| CmMADS39 | MIKCC | 246 | 27840.66 | 6.35 | 89.59 | -0.643 | 43.54 |
| CmMADS40 | MIKCC | 253 | 28963.06 | 7.66 | 80.95 | -0.773 | 59.52 |
| CmMADS41 | MIKCC | 314 | 35830.12 | 9.14 | 85.35 | -0.540 | 47.91 |
| CmMADS42 | MIKC* | 337 | 38123.77 | 5.20 | 74.69 | -0.624 | 62.24 |
| CmMADS43 | MIKC* | 230 | 26350.01 | 6.98 | 84.30 | -0.583 | 66.29 |
| CmMADS44 | MIKC* | 232 | 26173.76 | 6.25 | 84.87 | -0.584 | 59.92 |
| CmMADS45 | MIKC* | 540 | 61665.78 | 5.94 | 77.65 | -0.574 | 48.46 |
| CmMADS46 | MIKC* | 395 | 44844.68 | 7.66 | 76.05 | -0.599 | 49.81 |
| CmMADS47 | MIKCC | 196 | 22361.27 | 9.16 | 77.09 | -0.807 | 42.81 |
| CmMADS48 | MIKCC | 209 | 24488.94 | 6.62 | 83.97 | -0.828 | 61.86 |
| CmMADS49 | MIKCC | 225 | 26139.55 | 9.35 | 85.38 | -0.804 | 41.01 |
| CmMADS50 | MIKCC | 213 | 24388.17 | 9.25 | 101.08 | -0.488 | 52.81 |
| CmMADS51 | MIKCC | 265 | 30012.28 | 6.27 | 91.58 | -0.518 | 56.00 |
| CmMADS52 | MIKCC | 223 | 25229.90 | 5.80 | 89.60 | -0.617 | 50.61 |
| CmMADS53 | MIKCC | 222 | 25360.60 | 6.27 | 85.63 | -0.787 | 59.08 |
Phylogenetic Analysis of MADS-box Proteins
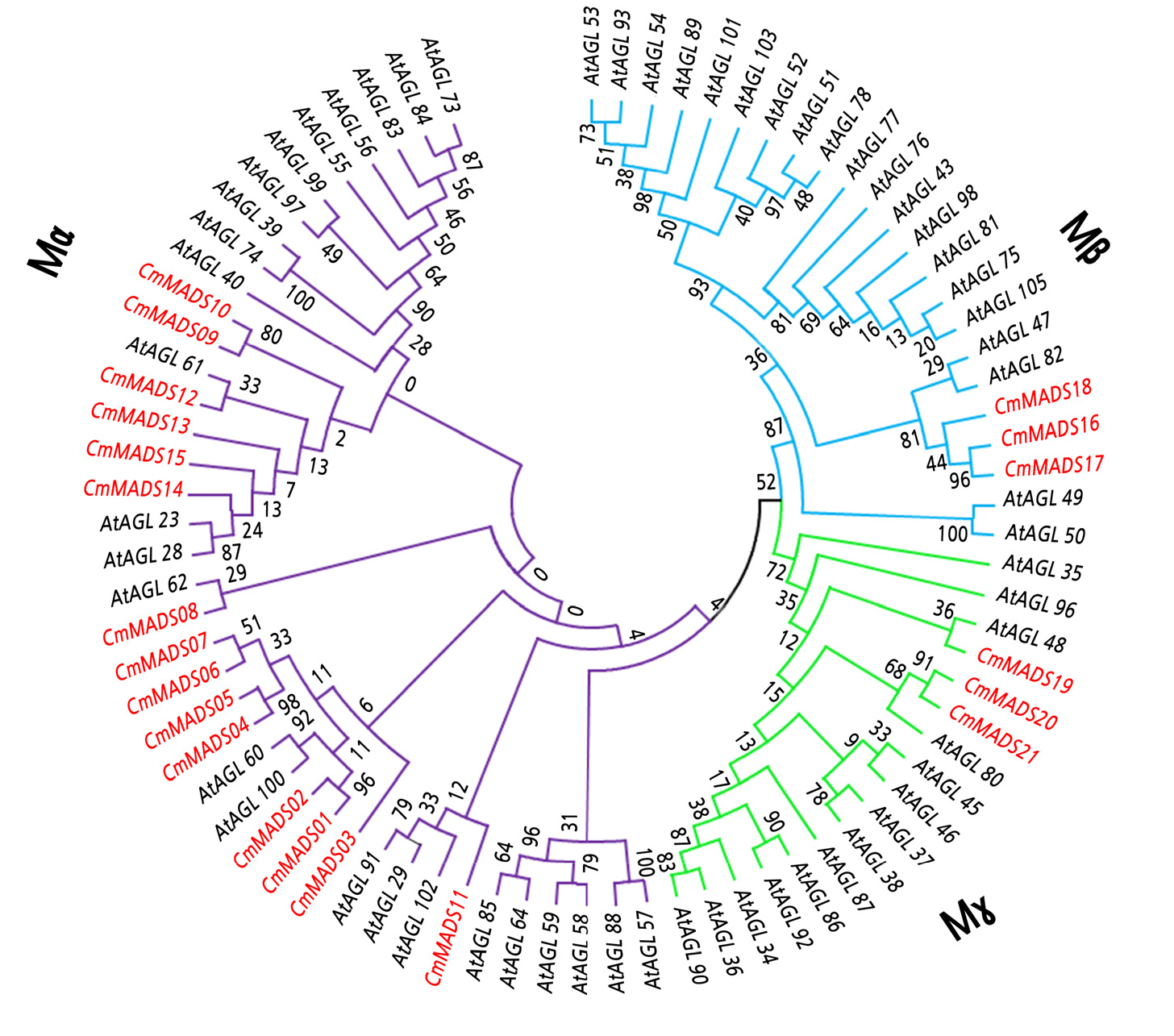
Type I and type II MADS-box protein phylogenetic trees were constructed by multiple sequence alignments of A. thaliana and the C. mollissimaMADS-box protein sequences. Type I proteins clustered into Mα, Mβ, and Mγ groups (Fig. 2). The Mα group had 15 genes, whereas the Mβ and Mγ groups each contained only 3 genes. Based on the phylogenetic trees, most type I genes were present in specific monophyletic lineages. For example, we observed A. thaliana homologs (AtAGL82 and AtAGL47) for three C. mollissima genes (CmMADS16, CmMADS17, and CmMADS18) of the Mβ group. However, there were no homologous genes in A. thaliana for four genes (CmMADS04, CmMADS05, CmMADS06, and CmMADS07) of the Mα group. Type II genes clustered into the MIKCC and MIKC* groups, with MIKCC containing 12 subgroups (Fig. 3). Type II subfamilies of Chinese chestnut contained similar subgroups based the phylogenetic trees; the exception was subgroup TM8 because there were no members of this subgroup in the A. thaliana genome (Heijmans et al., 2012). Therefore, we used the sequences of P. trichocarpaPtTM8 and P. persicaPpeTM8 for phylogenetic analysis and found CmMADS38 to be a TM8 member in Chinese chestnut, homologous to the genes in P. trichocarpa and P. persica (Fig. 3). To further investigate the CmMADS38 relationship, we created phylogenetic trees of TM8 genes from other plant species. CmMADS38 clearly belongs to a lineage of TM8 from Chinese hickory (Carya cathayensis), walnut (Juglans regia), poplar (P. trichocarpa), and cucumber (Cucumis sativus). Additionally, we identified three Chinese chestnut homologs each for the A. thaliana, AP3, and SVP subgroups; five for the AG subgroup and MIKC* group; four for the SEP subgroup; and two for the AGL6, AGL15, SOC1, and AP1/FUL subgroups. The AGL12, AGL17, and FLC subgroups contain only one homolog. However, no members of the BS subgroup exist in Chinese chestnut, represented by AtAGL63 and AtAGL32 in A. thaliana.
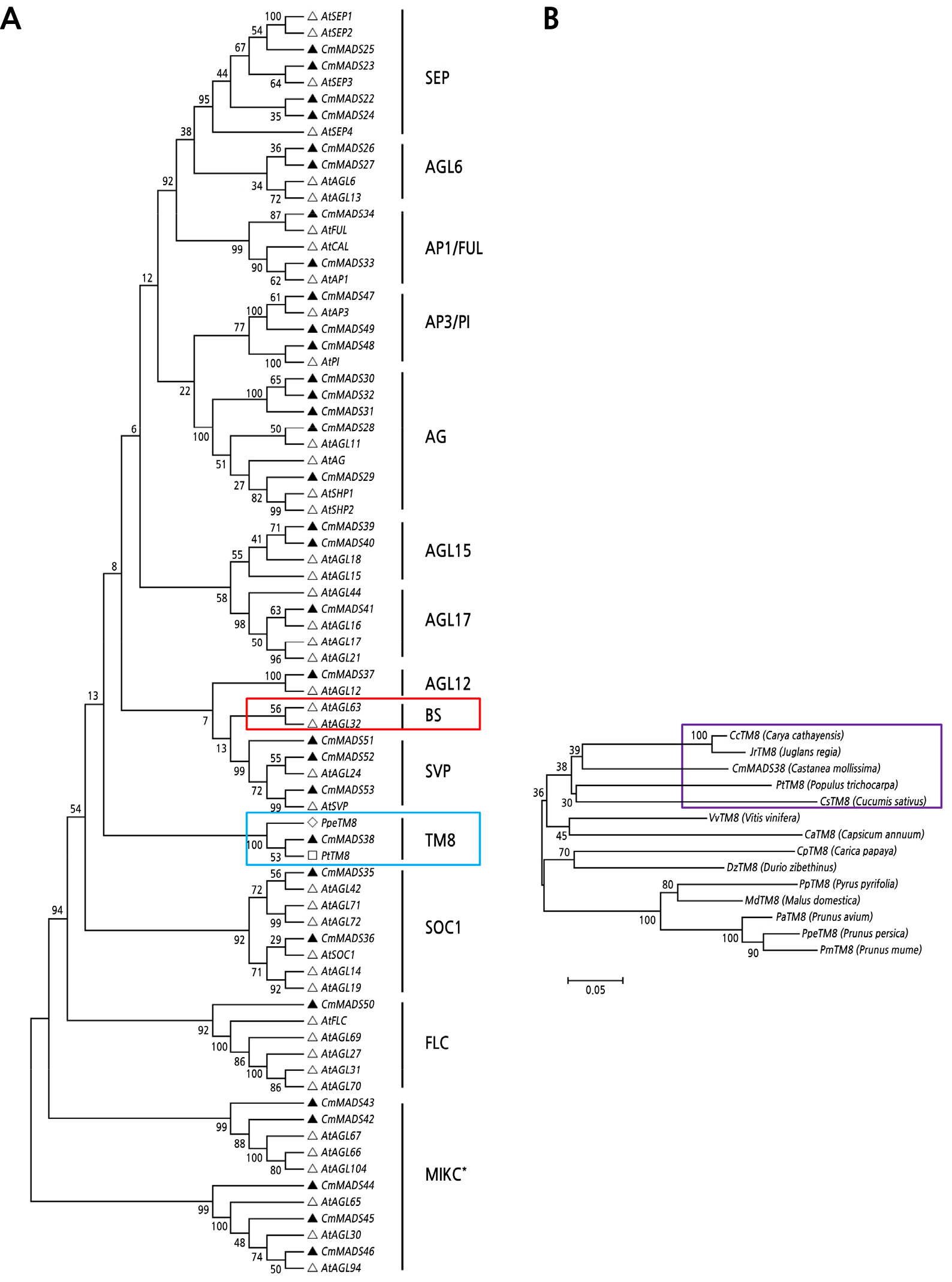
Fig. 3.
Phylogenetic tree of type II MADS-box genes in Chinese chestnut. (A) Phylogenetic tree of type II MADS-box genes in Chinese chestnut and A. thaliana; there were no members of the TM8 subgroup in the A. thaliana genome, so the sequences of poplar PtTM8 (Accession ID: XP_002321711.1) and peach PpeTM8 (Accession ID: XP_007211026.2) were downloaded for phylogenetic analysis. (B) Phylogenetic tree of TM8 subgroup genes in Chinese chestnut and several other species.
Conserved Motifs of MADS-box Proteins
Ten conserved motifs were identified from the 53 MADS-box genes of Chinese chestnut (Fig. 4). Among these motifs, motifs 1 and 2, encoding the MADS domain, were found in all Chinese chestnut CmMADS proteins except for the two Mα-group proteins (CmMADS06 and CmMADS07), which do not contain motif 1. Thus, motif 2 is the most conserved motif in the MADS domain. Motif 3, which denotes the K domain, was found in all MIKCC- and MIKC*-group proteins. However, part of the K domain (motif 3) is also present in CmMADS20 and CmMADS21, which are Mγ-group proteins. The type I clade contained unique motifs, and similar motif compositions are generally shared among proteins of this group (Fig. 4). Comparatively fewer instances of a conserved motif 4, representative of the I domain, were detected among MIKCC- and MIKC*-group proteins, except for CmMADS44. Some type I clade proteins also contain I domains. Finally, motif 10, representing the C-terminal domain of CmMADS proteins, is weakly conserved and present only in some MIKCC group proteins in Chinese chestnut.
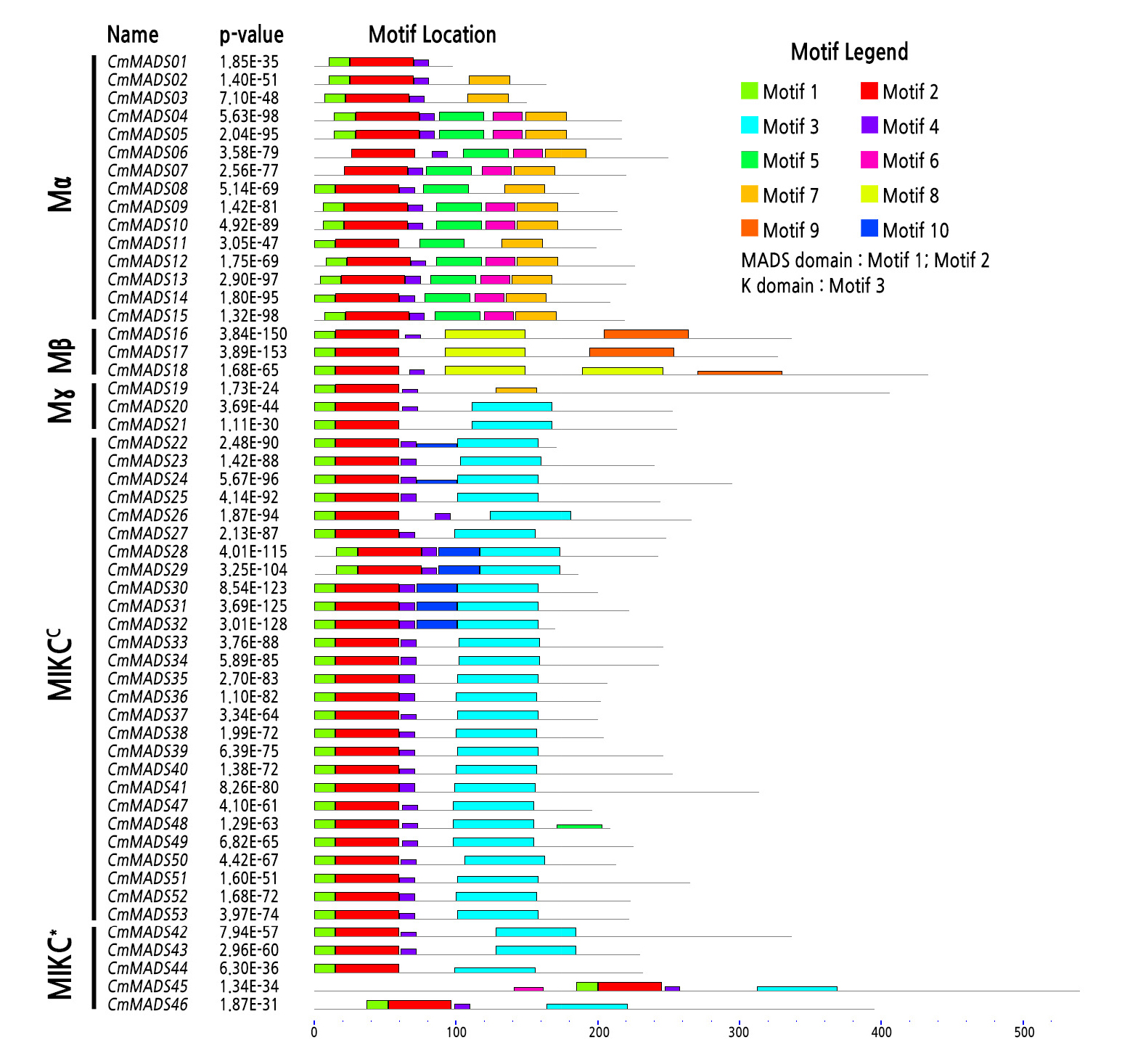
Fig. 4.
Motif composition of MADS-box proteins in Chinese chestnut. Each motif is represented by a colored block. The lengths and positions of the blocks correspond to the lengths and positions of motifs in the individual protein sequences. The height of each block is proportional to its -log (p-value), truncated at the height corresponding to a motif with a p-value of 1e-10. The gene names and combined p-value are shown on the left side of the figure. The scale pertains to the lengths of the proteins as well as the motifs.
Expression Analysis of CmMADS Genes in Reproductive Organs
Expression profiles of CmMADS genes were analyzed in organs from Chinese chestnut, anddiverse levels were observed (Fig. 5). Overall, type I genes had low expression and the expression of 8 of these genes was absent. According to the heat map of gene expression, CmMADS genes were expressed in reproductive organs (mixed floral bud, female flower, and staminate catkin), vegetative organs (leaf), and cotyledons of Chinese chestnut. AG, TM8, AP1/FUL, AGL6, SEP, SOC1, SVP, and FLC subgroups, and MIKC* group genes exhibited higher expression levels than did other subfamily genes in the mixed floral bud and female flower, and AG, AP1/FUL, AGL6, SEP, and SOC1 subfamily genes were highly expressed in the stigma and ovary. However, the expression of the AP3/PI subgroup genes CmMADS48 and CmMADS49 was also high in the stigma and ovary. CmMADS38, a TM8 subfamily gene, showed higher expression levels in the ovary than in other organs. Moreover, AG, AP3/PI, TM8, AP1/FUL, AGL6, SEP, SOC1, SVP, and FLC subgroup, and MIKC* group genes were more highly expressed than other subfamily genes in the mixed floral bud and female flower. In addition, the expression of AG, AP3/PI, AP1/FUL, AGL6, SEP, SOC1, and SVP subgroup, and MIKC* group genes was high in the floral cluster. In the stamen, AG, AP3/PI, and SEP subgroup and MIKC* group gene expression, especially CmMADS48 and CmMADS46, was higher than that of other MADS-box genes. SVP, SOC1, and AP1/FUL subgroup genes appear to regulate leaf development and CmMADS11, CmMADS34, CmMADS45, and CmMADS50 appear to be involved in cotyledon development.
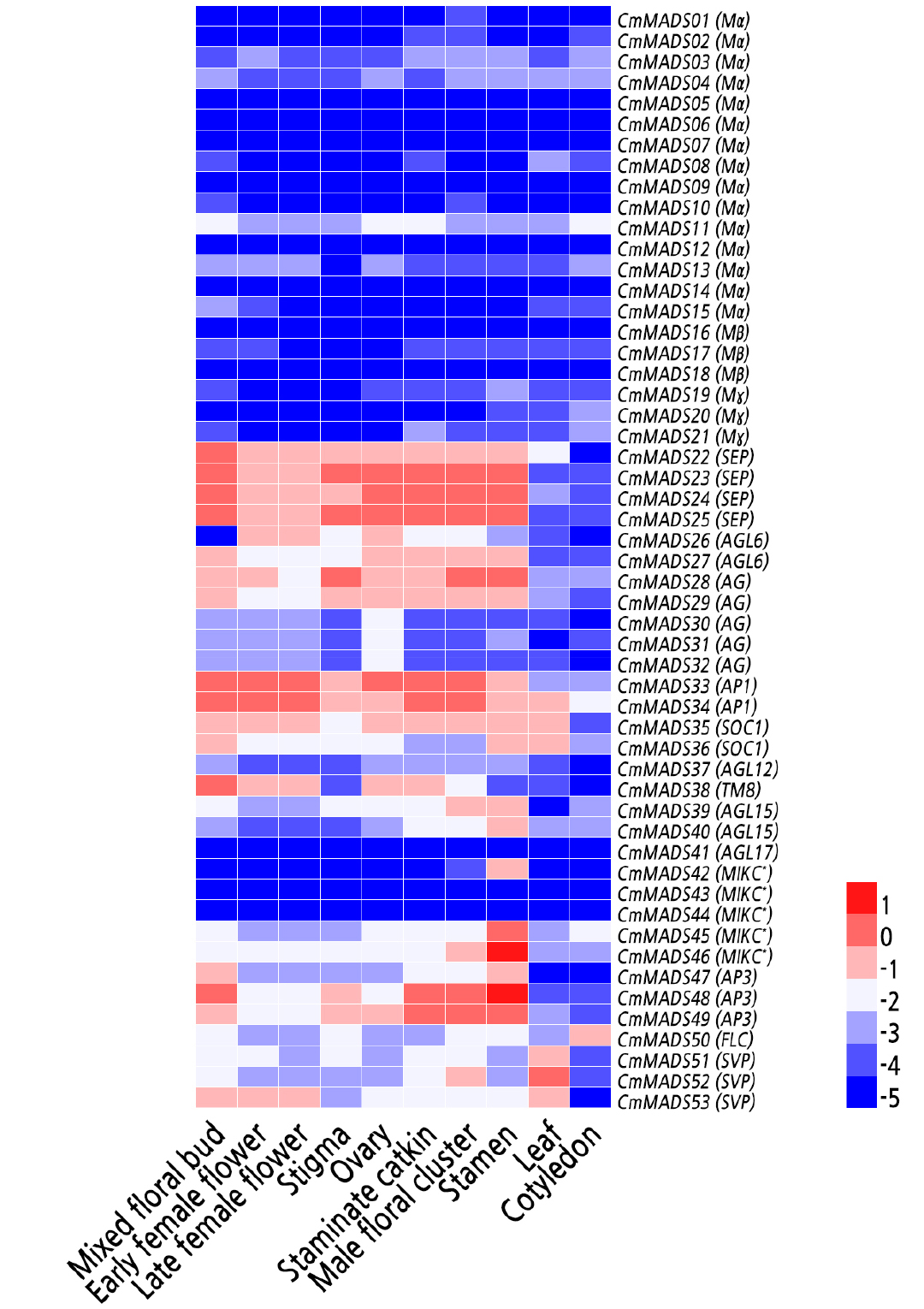
Fig. 5.
Expression patterns of CmMADS genes in different organs of Chinese chestnut. The heat map was generated based on the mean expression values of the qRT-PCR data from different samples. The mean relative expression values are listed in Suppl. Table 2s, and normalized for the heat map using the HemI 1.0.3.3 tool. CmActin was used as the control. Red and blue represent higher and lower expression, respectively.
To further investigate the roles of CmMADS genes in reproductive organs, the expression of MADS-box genes was analyzed in the inflorescence primordium early differentiation stage (B1), inflorescence primordium later differentiation stage (B2), floral clusters bract primordium formation stage (B3), and floral clusters primordia shaped formation stage (B4). Based on the heat map (Fig. 5), 16 CmMADS genes were highly expressed in floral organs and their expression decreased continuously during mixed floral bud development, though these genes were highly expressed in the B1 stage (Fig. 6).
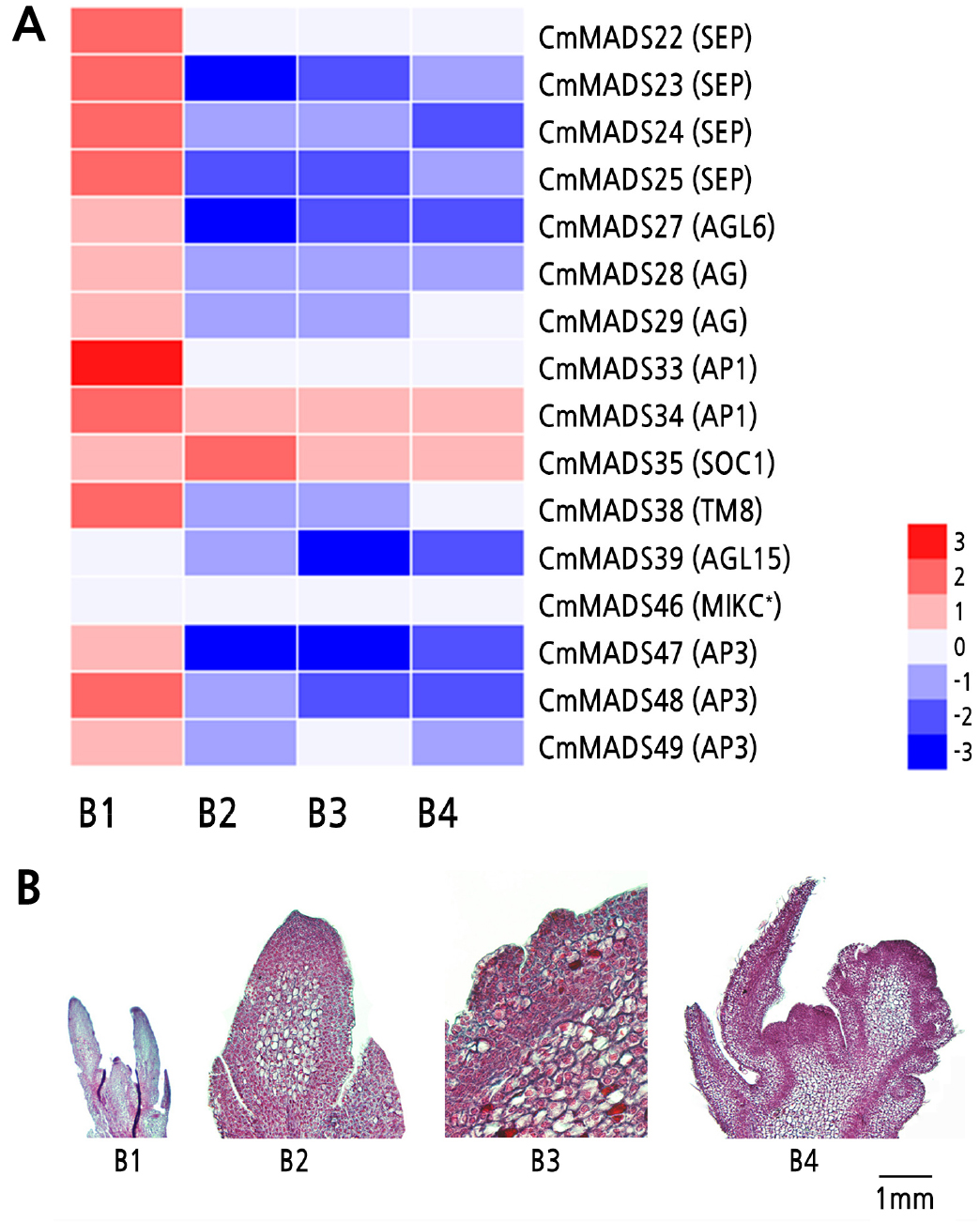
Fig. 6.
Expression patterns of 16 CmMADS genes during mixed floral bud development. (A) Heat map showing the expression of 16 MADS-box genes during mixed floral bud development. The mean relative expression values are listed in Suppl. Table 3s, and normalized for the heat map using the HemI 1.0.3.3 tool. CmActin was used as the control, (B) The morphology of mixed floral bud development stages B1-B4 in Chinese chestnut; B1-B4 represent inflorescence primordium early differentiation stage, inflorescence primordium later differentiation stage, floral clusters bract primordium formation stage, and floral clusters primordia shaped formation stage, respectively.
In addition, we further examined genes with high expression levels in floral organs but not in vegetative organs (Table 2). SEP, AGL6, AG, AP1, TM8, AGL15, and AP3/PI subgroup and MIKC* group genes appear to be involved in female flower and staminate catkin development, with CmMADS31 having high expression levels in the female flower. SEP, AGL6, AG, AP1, AGL15, and AP3/PI subgroup and MIKC* group genes appear to participate in stigma development. Based on their expression, SEP, AGL6, AG, AP1, TM8, AGL15, and AP3/PI subgroup and MIKC* group genes are involved in ovary development, and CmMADS30 and CmMADS32 showed high expression levels in the ovary. Development of the male floral cluster appears to involve SEP, AGL6, AG, AP1, AGL12, TM8, AGL15, and AP3/PI subgroup and MIKC* group genes, and stamen development appears to involve SEP, AGL6, AG, AP1, AGL15, and AP3/PI subgroup and MIKC* group genes, with CmMADS42 showing high expression levels in the stamen of Chinese chestnut.
Table 2.
List of CmMADS genes highly expressed in Chinese chestnut flower organs
Discussion
In this study, we identified 53 MADS-box genes in Chinese chestnut. The number of MADS-box genes in Chinese chestnut was lower than that in P. trichocarpa (101) (Leseberg et al., 2006), P. persica (79) (Wells et al., 2015), M. domestica (146) (Tian et al., 2015), V. vinifera (Grimplet et al., 2016), A. thaliana (108) (Parenicová et al., 2003), and Oryza sativa (76) (Arora et al., 2007) but greater than that in C. sativus (43) (Hu and Liu, 2012) and Z. jujuba (52) (Zhang et al., 2017). The number of type I subfamilies in Chinese chestnut, Z. jujube, and C. sativus was significantly lower than that in other species (Table 3). Gene family expansion and contraction may be among the primary reasons for functional changes. Phylogenetic analysis suggested that tandem duplication was responsible for the expansion and contraction of MADS-box genes in Chinese chestnut. In A. thaliana, 5 type I genes (AGL80, PHERES1, DIANA, AGL62, and AGL23) are reportedly involved in the development of the female gamete and early seed, but the functions of the other genes are still unknown (Bemer et al., 2010). Type I genes CmMADS13 and CmMADS15 hadhigh expression levels in the mixed bud and female flower compared to the male flower in Chinese chestnut. Therefore, CmMADS13 and CmMADS15 may be involved in female flower development in Chinese chestnut, which remains to be functionally explored in future studies.
Table 3.
Numbers of MADS-box genes in Castanea mollissima, Populus trichocarpa, Prunus persica, Malus domestica, Vitis vinifera, Ziziphus jujuba, Cucumis sativus, Arabidopsis thaliana, Oryza sativa
According to the phylogenetic analysis of MADS-box genes, Chinese chestnut contains the TM8 subgroup, which does not exist in A. thaliana, whereas the BS subgroup found in A. thaliana is absent in Chinese chestnut. CmMADS38, a TM8 subgroup gene, was more highly expressed in flower organs than in the leaf and cotyledon, with the highest and lowest amount of transcript among all floral organs in the ovary and stamen, respectively. TM8 was first isolated from the arrested floral meristem of the anantha mutant of tomato (Solanum lycopersicum) but not pursued for more than twenty years (Daminato et al., 2014); thus, TM8 was called the dark side of floral development (Heijmans et al., 2012). Since then, TM8 has been detected in Antirrhinum majus, P. trichocarpa, and V. vinifera, though no TM8-clade genes were functionally characterized in any species until after 2012 (Heijmans et al., 2012). The expression level of TM8 is higher in S. lycopersicum flowers than in leaves and cotyledons, and the greatest amount of transcript is found in petals, followed by anthers. TM8-overexpressing S. lycopersicum plants exhibit anomalous stamens with low-viability pollen, anomalous ovaries and seeds, and altered expression of several floral identity genes belonging to the B-, C-, and E-function groups (Daminato et al., 2014). Therefore, we deduce that CmMADS38 may regulate ovary development and contribute to the low fertility rate and embryo abortion in the ovary of Chinese chestnut.
To date, most studies to identify the function of MADS-box genes have been conducted in herbaceous and woody plants, revealing the diverse functions of MADS proteins (Hu and Liu, 2012; Wells et al., 2015; Tian et al., 2015; Zhang et al., 2017). To gain insight into Chinese chestnut MADS-box gene expression patterns, we analyzed gene expression by qRT-PCR in 10 different tissues. The results revealed higher expression levels in floral organs than in leaves and cotyledons of close orthologs of SEP, AGL6, AG, AP1/FUL, AGL15, AP3, and MIKC* genes in Chinese chestnut, which indicates an association with the development of reproductive organs. Although Chinese chestnut flowers develop two whorls of organs, the expression model of MADS-box genes seems to be different from the ABCDE model of four-whorled flowers in A. thaliana (Thomson et al., 2017). According to the expression of MADS-box genes in Chinese chestnut, the male flower may be specified by the ABE function genes, while the female flower may be specified by the ACE function genes. This is similar to Quercus suber, a monoecious species in Fagaceae, in which the BCE combinatorial complex is involved in male flower development, whereas the CE complex is involved in female flower development (Sobral and Costa, 2017). In Chinese chestnut, SEP, AP1/FUL, SOC1, and MIKC* genes were highly expressed in mixed floral bud development, especially in the inflorescence primordium early differentiation stage (B1), suggesting that these genes may affect the transition from vegetative to reproductive development. In addition, the AP1/FUL subfamily genes are involved in the development of the sepal and petal (Thomson et al., 2017). The AP1/FUL subfamily genes in Chinese chestnut are involved in male and female flower development; therefore, it is possible that flower organs do not have the sepal and petal in Chinese chestnut. In addition, we identified five MIKC* group genes in Chinese chestnut. CmMADS42, CmMADS45, and CmMADS46 were highly expressed in the stamen in Chinese chestnut. MIKC* MADS proteins play an essential role in regulation during pollen development in A. thaliana (Verelst et al., 2007a, 2007b). In Chinese chestnut, male flowers far outnumber female flowers and the pollen germination rate is low (Feng et al., 2011). CmMADS42 has the lowest pI value, showing a high expression level only in the stamen. We deduced that CmMADS42 may regulate pollen development in Chinese chestnut. Taken together, the results of this study provide valuable information for future studies of the regulation of CmMADS genes in Chinese chestnut flower development.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20210043_Table_1s.pdf
Primer sequences used for qRT-PCR
- HORT_20210043_Table_2s.pdf
The mean relative expression value of the Chinese chestnut MADS-box genes in different organs
- HORT_20210043_Table_3s.pdf
The mean relative expression value of the 16 MADS-box genes during mixed floral bud development