Introduction
Materials and Methods
L. cylindrica Genome Source
Plant Materials
Development of Polymorphic SSR Markers
SSR Genotyping and Polymorphic Analysis
Results
Characterization of Polymorphic SSRs in L. cylindricaL. cylindrica
Validation of Polymorphic SSR Markers in Luffa Samples
Application of Polymorphic SSR Markers in Genetic Diversity Analysis
Discussion
Introduction
The genus Luffa comprises two cultivated species, Luffa cylindrica (sponge gourd) and L.acutangula (ridged gourd). L. cylindrica is a common vegetable crop mostly cultivated in India, China, and Southeast Asia (Filipowicz et al., 2014). It has high nutritional value and a delicious taste. Specifically, L. cylindrica is rich in carbohydrates and various mineral elements. It also contains various biologically active substances such as glycosides, alkaloids, and flavonoids (Yan et al., 2011). L. cylindrica is also a medicinal plant with special medicinal value, including antimicrobial and anti-cancer properties (Sharma et al., 2019). Compared with other cucurbit crops, research on the genetics and breeding of L. cylindrica has lagged. Until now, most of the new varieties have been improved by traditional methods (Dai et al., 2016).
High-throughput sequencing has accelerated large-scale development of crop molecular markers. However, screening polymorphic molecular markers is still an expensive approach. Simple sequence repeats (SSRs), also known as microsatellite markers, have high stability and polymorphism and thus have become one of the most applied mainstream molecular markers (Ellegren, 2004; Wang et al., 2015). Researchers have mainly developed SSR markers of L. cylindrica through transcriptome sequencing prior to whole-genome sequencing of the species (Wu et al., 2014; Zhu et al., 2016b). With the completion of whole-genome sequencing of L. cylindrica (Wu et al., 2020; Zhang et al., 2020; Pootakham et al., 2021), 284 966 SSR markers in the whole genome were developed (Qiao et al., 2021). However, large-scale development of polymorphic SSR markers in L. cylindrica by genome prediction remains poorly explored.
In this study, two genomic sequences of L. cylindrica ‘SG2019’(Zhang et al., 2020) and ‘SO3’ (Pootakham et al., 2021), were utilized to develop polymorphic SSR markers for L. cylindrica. SSR loci were identified at the same site with different repeats between the two genomes. Furthermore, 40 pairs of predicted polymorphic SSR primers were randomly selected for SSR genotyping of 24 Luffa samples, and their high polymorphic rate was verified. The polymorphic SSR markers developed in this study can improve the efficiency of SSR markers and accelerate the process of marker-assisted selection in L. cylindrica.
Materials and Methods
L. cylindrica Genome Source
Polymorphic SSR markers were developed based on two published genomes of L. cylindrica. The two genomes were ‘SG2019’ (Zhang et al., 2020) from cngbdb (https://db.cngb.org/), accession number cnp0000780, and ‘SO3’ (Pootakham et al., 2021) from NCBI (https://www.ncbi.nlm.nih.gov/), accession number GCA_ 012295205.1.
Plant Materials
A total of 24 Luffa samples (Table 1) were used to verify the polymorphism of developed SSR markers. The 24 Luffa samples included landraces and commercial cultivars of L. cylindrica, except for ‘Shengyou’, ‘Meizhou’ and ‘Shengyou × Pingguo F1’ (first generation of interspecific hybrid between L.acutangula ‘Shengyou’ and L. cylindrica ‘Pingguo’).
Table 1.
Basic information of 24 Luffa samples
Development of Polymorphic SSR Markers
MISA (http://pgrc.ipk-gatersleben.de/misa/) was used to search for SSR loci of the two L. cylindrica genomes ‘SG2019’ and ‘SO3’. The search parameters were: (1) the SSR length was larger than 22 bp; (2) the length of sequences on both sides of the SSR site was set to 150 bp; (3) motif repeat times ≥ 4; (4) The length of the motif was 2–6, and the corresponding SSR motif types were dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide.
Primer3 software was used to design primers for SSR loci found in the two genomes based on the following parameters: ‘prime_product_size_range = 80-160 80-240 80-300; prime_opt_size = 24; prime_min_size = 20; prime_max_size = 28; prime_opt_tm = 63; prime_min_tm = 60; prime_max_tm = 65’. After primer design, the upstream and downstream primer sequences of each SSR locus were aligned back to the reference genome of ‘SG2019’, and only the SSR loci specifically aligned to the primer sequences were retained.
Finally, using ‘SG2019’ as the reference genome, SSR loci with different motif repeats between ‘SG2019’ and ‘SO3’ were identified using the Mummer program (http://mummer.sourceforge.net/manual/).
SSR Genotyping and Polymorphic Analysis
The SSR reaction system and amplification were performed as described in a previous report (Cui et al., 2017). The amplification products were resolved in 6% polyacrylamide gels and stained with silver. When an SSR marker gave an amplification product on at least one of the samples, this indicated that the marker could be amplified. Based on the electrophoresis results, a clear band was marked as ‘1’ and an absence of a band at the same position was marked as ‘0’, forming a ‘0-1’ matrix. The data were converted into different formats using DataFormater software (Fan et al., 2016) as required. The polymorphism information content (PIC) of each SSR marker was calculated using Powermarker software (Liu and Muse, 2005). The cluster analysis was analyzed using NTSYS-pc software (http://www.appliedbiostat.com/ ntsyspc/ntsyspc.html), whereas the population structure was analyzed using structure software (Pritchard et al., 2000).
Results
Characterization of Polymorphic SSRs in L. cylindrica
A total of 21249 and 21303 SSR loci were identified in L. cylindrica genomes ‘SG2019’ and ‘SO3’, respectively. Then, taking the ‘SG2019’ genome as a reference, 4613 (21.71%) concomitant SSR loci and 2130 (10.02%) SSR loci with different motif repeats were found by comparing the SSR motif repeats in the two L. cylindrica genomes (Table 2, and Suppl. Table 1). Among the 2130 predicted polymorphic SSR loci, dinucleotides accounted for the largest proportion (1710; 80.25%), followed by trinucleotides (322; 15.12%). The number of tetranucleotides, pentanucleotides, and hexanucleotides was 40 (1.88%), 19 (0.89%), and 39 (1.83%), respectively. In addition, the 2130 polymorphic SSR loci contained 54 types of motif units, of which AT/AT and AAT/ATT accounted for 87.80% of the total number.
Table 2.
Polymorphic SSR motifs predicted from two L. cylindrica genomes
Validation of Polymorphic SSR Markers in Luffa Samples
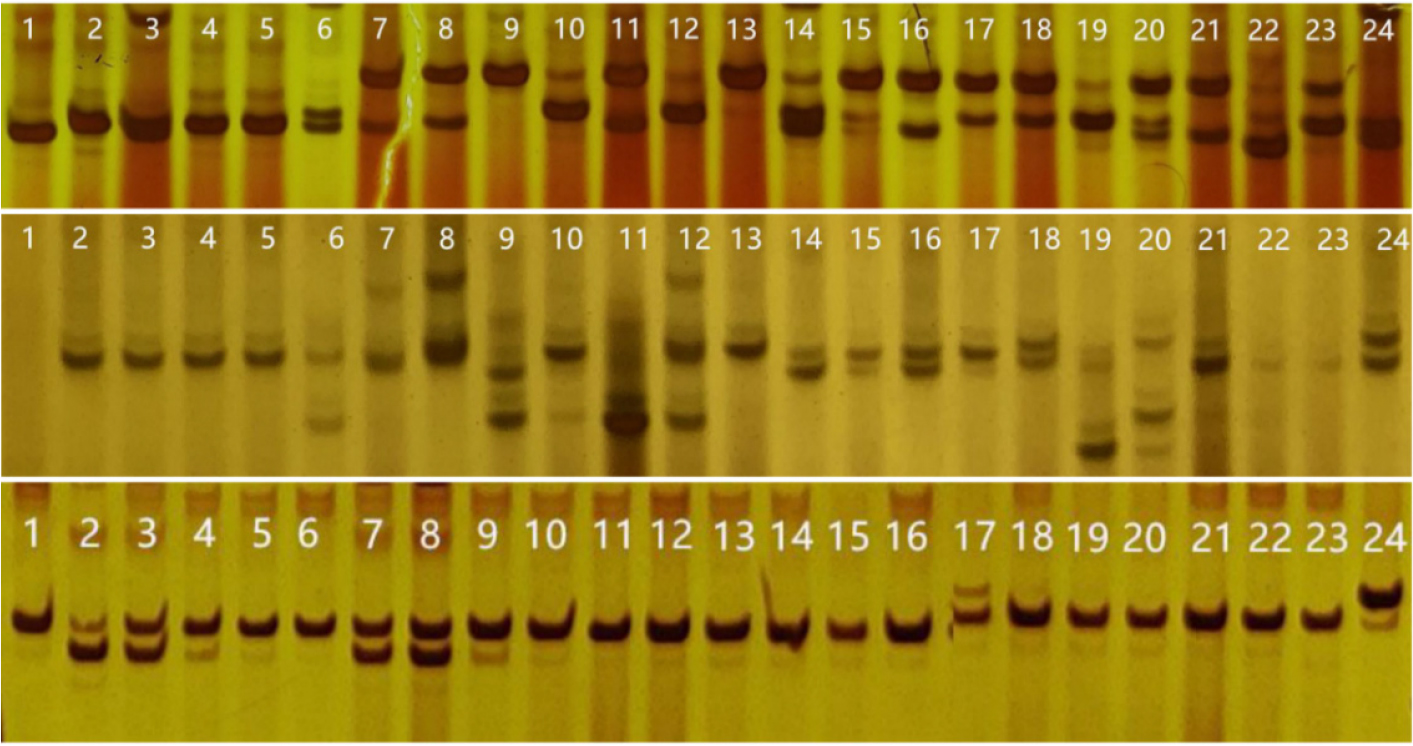
To validate the polymorphism of the newly developed polymorphic SSR markers, we randomly selected 40 SSR markers that were distributed on all chromosomes and contained different motif types to genotype the 24 Luffa samples. Amplification results showed that all 40 SSR markers bound to specific regions in some or all the 24 Luffa samples, with a 100% amplification rate. Furthermore, 32 out of the 40 SSR markers showed polymorphism in the 24 Luffa samples, with an 80% polymorphic rate. The 32 SSR markers amplified 2–5 alleles in the 24 Luffa samples, with an average of 3.16 (Table 3). The PIC values of the 32 SSR markers in the 24 Luffa samples ranged from 0.08 to 0.65, among which the PIC values of pLC0777, pLC0767, and pLC1223 were 0.65, 0.57, and 0.50, respectively, indicating a high level of polymorphism (Fig. 1).
Table 3.
Sequence information and polymorphism verification of 32 polymorphic SSR primers in Luffa
Application of Polymorphic SSR Markers in Genetic Diversity Analysis
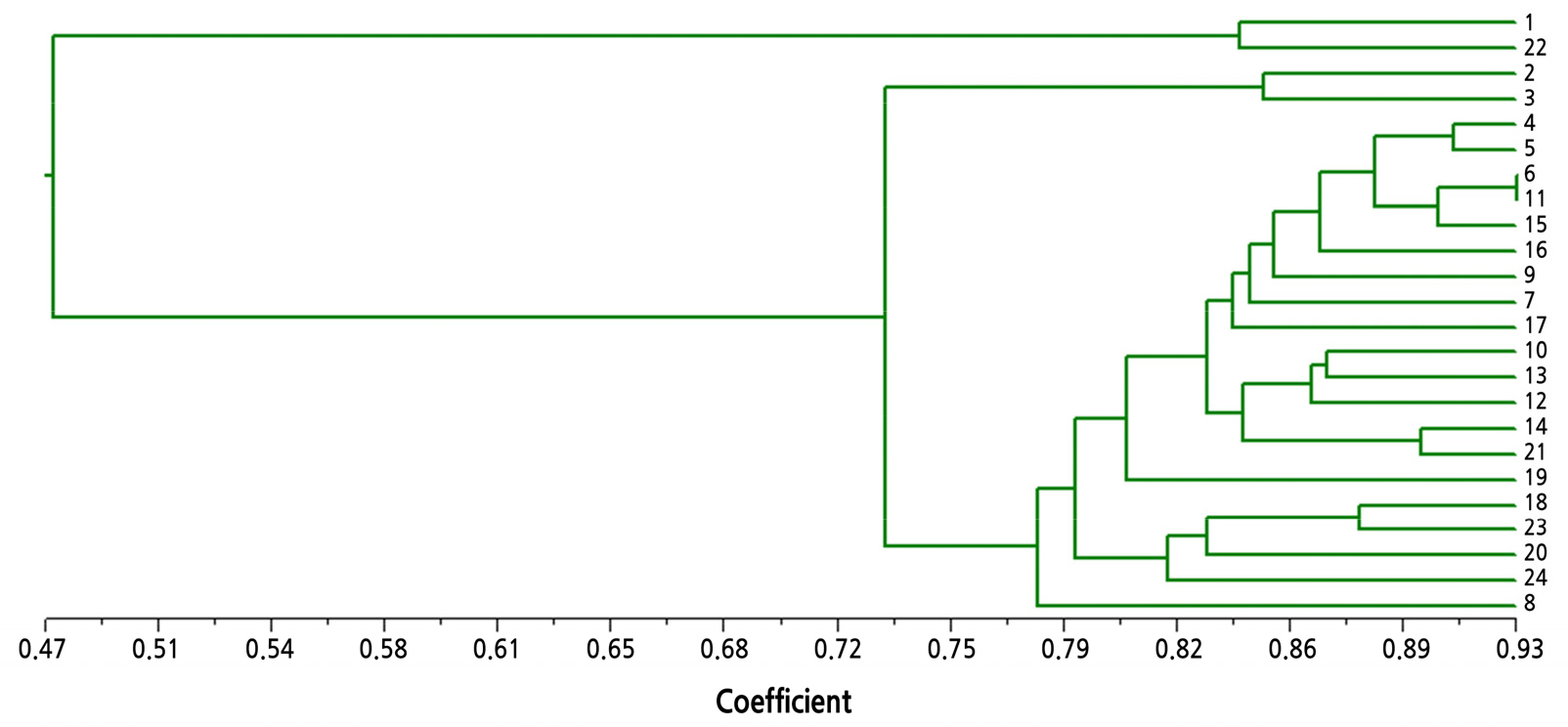
Cluster analysis was conducted on the 24 Luffa samples using 32 polymorphic SSR markers from L. cylindrica. The 24 Luffa samples were divided into two groups at a genetic similarity coefficient of 0.47 (Fig. 2).
Group Ⅰ included two L.acutangula samples, ‘Shengyou’ (No. 1) and ‘Meizhou’ (No. 22), whereas group II included 22 Luffa samples. At a genetic similarity coefficient of 0.80, the 22 Luffa samples in group II were divided into four subgroups: group II-1 included two samples, ‘Pingguo’ (No. 2) and ‘Shengyou × Pingguo F1’ (No. 3); group II-2 included one L. cylindrica sample, ‘Zaozhou No.2’ (No. 8); group II-3 included four L. cylindrica samples, ‘Yulin’ (No. 24), ‘Fengyou No. 3’ (No. 20), ‘Changfeng’ (No. 23), and ‘Juxing No.1’ (No. 18); and group II-4 included the remaining 15 L. cylindrica samples.
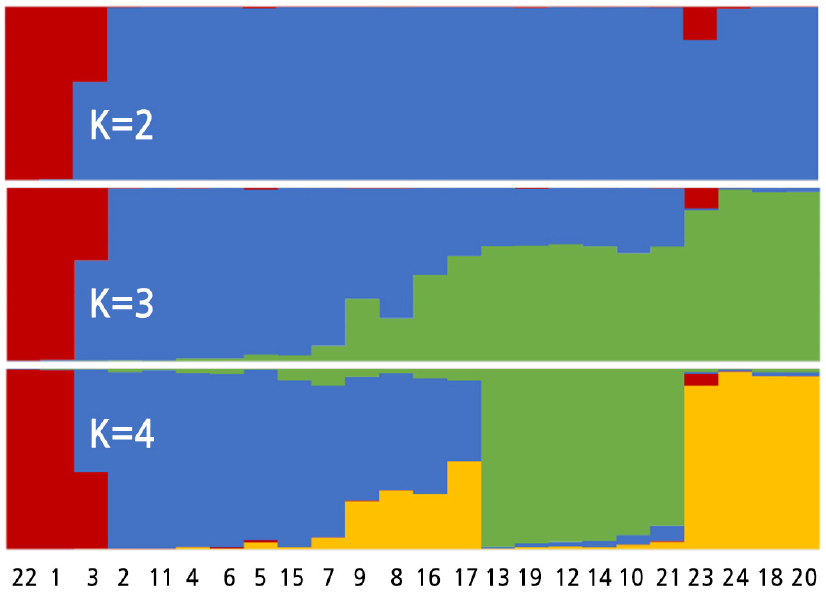
The population structure of the 24 Luffa samples was analyzed using structure software. The structure analysis results were consistent with the clustering results and specifically reflected the kinship of different samples (Fig. 3). For example, a population structure of K ranging from 2 to 4 indicated that the interspecific hybridization, ‘Shengyou × Pingguo F1’ (No. 3), came from two parents. The commercial cultivar L. cylindrica ‘Changfeng’ (No. 23) was more likely to contain a small fraction of genetic background from L.acutangula germplasm.
Discussion
Genome-wide SSR markers of L. cylindrica have recently been developed (Qiao et al., 2021). The availability of efficient and reliable polymorphic markers significantly reduces the screening costs and delays during the validation of large-scale SSR markers. There are few reports on the application of SSR markers in L. cylindrica. For example, there are a few reports on the application of SSR markers for the analysis of genetic diversity of germplasm resources (Liu et al., 2010), the construction of genetic maps and mapping of flowering time (Wu et al., 2016), and the identification of hybrid purity (Zhu et al., 2020).
In this study, 2130 SSR markers showing polymorphism between L. cylindrica genomes ‘SG2019’ and ‘SO3’ were obtained by comparing their genomic sequences. The number of dinucleotide and trinucleotide SSR loci in the genome of L. cylindrica was predicted to be 54363 and 28972, respectively (Qiao et al., 2021). However, the proportion of dinucleotide SSR loci was 80.28%, far exceeding the proportion of trinucleotide SSR loci (15.12%), indicating that dinucleotide SSR loci are the main type of polymorphism in the L. cylindrica genome. The 2130 polymorphic SSR markers contained 54 kinds of motif units, which was much lower than the 211 kinds in the whole genome of L. cylindrica (Qiao et al., 2021), indicating that the polymorphic SSR markers of L. cylindrica are skewed towards some types of motif units. Among the polymorphic SSR markers of L. cylindrica, AT/AT and AAT/ATT had the largest number of motif units, consistent with the L. cylindrica genome. In this study, the designed SSR primer sequences were aligned back to the reference genome, and those that aligned uniquely were retained to ensure the amplification of the SSR markers. A total of 40 SSR markers were amplified in 24 Luffa samples, with an amplification rate of 100%. Among the 40 SSR markers verified, 32 were polymorphic with a polymorphic rate of 80%, which was higher than that reported previously (Wu et al., 2014; Zhu et al., 2016b; Qiao et al., 2021).
Based on the standard of polymorphism level (Botstein et al., 1980), only three out of the 32 polymorphic SSR markers verified in this study were highly informative (PIC ≥ 0.50), while the other 28 were reasonably informative (0.25 ≤ PIC < 0.50) or slightly informative (PIC < 0.25). Notably, the difference in polymorphic level between these markers may be largely dependent on the genetic background of Luffa samples amplified. Although we developed a set of polymorphic SSR markers based on two L. cylindrica genomes, the high polymorphic rate observed in the Luffa population provides a reliable guarantee for their utilization. Cluster analysis of 24 Luffa samples showed that L. cylindrica and L.acutangula were separated into two large groups, consistent with the clustering results obtained using other molecular markers such as SRAP (Cui et al., 2012; Wu et al., 2015; Zhu et al., 2016a) and ISSR (Ye et al., 2017; Guo et al., 2020). The huge genetic difference between L. cylindrica and L.acutangula provides great potential for continuous improvement of the two Luffa species. Population structure analysis showed that interspecific hybridization between L. cylindrica and L.acutangula such as ‘Shengyou × Pingguo F1’ (No. 3) or genetic introgression such as ‘Changfeng’ (No. 23) could be an effective approach to genetic improvement of the Luffa species. Among the L. cylindrica samples, except that the only one sample with round-shaped fruit ‘Pingguo’ (No. 2) and the only one sample with wrinkled fruit skin ‘Zaozhou No.2’ (No. 8) was assigned to group II-1 and group II-2, respectively, the remaining samples showed a relatively close genetic distance, suggesting that fruit shape and skin were typical grouping characteristics. In conclusion, the polymorphic SSR markers developed from two L. cylindrica genomes in this study have a high polymorphic rate in Luffa species and can be used as primary candidate markers for genotyping Luffa species in the future.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20220049_Table_1s.xlsx
List of 2130 polymorphic SSR markers predicted from L. cylindrica