Introduction
Materials and Methods
Plant Materials and Genomic DNA Extraction
Mining of Polymorphic Plastid InDel loci and Primer Design
Genotyping by M13-tailed PCR and Data Analysis
Results and Discussion
Development of Polymorphic InDel Markers from Citrus Plastid Genomes
Discrimination of lemons from Other Citrus Groups using Polymorphic InDel Markers
Introduction
Lemon [Citrus limon (L.) Burm. F.] is considered to be native to the Southern part of the Himalayas in India or Southern China and probably Upper Burma (Nicolosi, 2007). Recent molecular studies suggest that lemons (yellow lemon types) resulted from the interspecific hybridization between sour orange (C. aurantium L.) and citron (C. medica L.) (Nicolosi et al., 2000; Curk et al., 2015; Curk et al., 2016). Sour orange itself is believed to be a hybrid between pummelo (C. maxima Burm.) and mandarin (C. reticulata Blanco) (Wu et al., 2018). However, volkamer and rough lemons are the products of hybridization between mandarin and sour orange (Curk et al., 2016). Lemons are generally divided into three types; ‘Eureka’, ‘Lisbon’, and others (Park et al., 2020).
Lemons are cultivated in Mediterranean, subtropical, and intertropical climates worldwide. However, only a few lemon varieties are cultivated worldwide for the production of fresh fruits and essential oils or for use as rootstocks, although substantial genetic diversity exists in this citrus group (Curk et al., 2016). In Korea, lemons have been commercially cultivated since the year 2000 and around 100 farms currently produce lemons from a cultivated area of 30 hectares. This cultivation area is expected to increase due to continuous market demands and farm profitability in Korea. Eureka-type lemons, such as ‘Allen’, ‘Frost’, and ‘Cook’, are mainly cultivated in Korea. It is difficult to clearly identify lemon varieties, even though various types of lemons, including ‘Lisbon’ and ‘Meyer’, are also assumed to be introduced into Korea. Microsatellite markers would be a robust molecular tool for identifying citrus cultivars including lemons (Woo et al., 2020).
Trifoliate orange [Poncirus trifoliata (L.) Raf.] rootstocks have been widely used in the citrus industry, including in Korea, due to several merits including a compact canopy, fruit quality, cold hardiness, and resistance to pathogens and pests. The rootstock trifoliate is quite suitable for satsuma mandarin (C. unshiu Marc.) that is the most prevalent citrus in Korea, accounting for approximately 87% of the citrus production in the year 2019. Recent trends for diversification of commercial citrus cultivars resulted in increased demand for more rootstock varieties (Kawase et al., 1987; Donadio et al., 2019). Various rootstock varieties such as lemons and their hybrids, lime, citrange, citrumelo, and Cleopatra mandarin were developed (URL https://citrusvariety.ucr.edu/citrus/rootstocks.html) and have been used in the citrus industry due to the drawbacks of trifoliate orange rootstocks, including incompatibility with several cultivars, virus problems, and nonvigorous growth (Kawase et al., 1987). In addition to these drawbacks, the seed germination rate of trifoliate orange rapidly decreases after May, whereas lemon seeds maintain a high germination rate throughout the year, guaranteeing high seedling yield and quality for rootstock production. However, a critical problem is that it is very hard to identify the genetic origin of new shoots when the micro-grafted scions are germinated on lemon rootstocks.
Several types of DNA-based molecular markers, such as simple sequence repeats (SSRs), single nucleotide polymorphisms (SNPs), and Insertions/Deletions (InDels) have been developed and used for the identification of nucellar/zygotic individuals (Woo et al., 2019), the efficiency test of hybrid embryo rescue (Kim et al., 2020), species/cultivar identification (Woo et al., 2020; Jin et al., 2020), and phylogenetic origin analysis in citrus (Curk et al., 2016). SSRs and InDel markers from the mitochondria and/or chloroplast genomes are widely used for the identification and authentication of crop cultivars (Curk et al., 2016; Park et al., 2017; Park et al., 2020; Roy et al., 2020).
In this study, we developed reliable, lemon-specific, multi-locus InDel markers from the comparative analysis of citrus plastid genomes including lemon, pummelo, sweet orange, and mandarin. This set of markers would be a useful molecular genetic tool for the rapid and clear discrimination of micro-grafted scion cultivars and lemon rootstocks during the production of virus-free citrus trees and for the guarantee of nursery stock quality in the citrus industry. The InDel markers from the maternally inherited plastid genome can also be used to analyze the phylogenetic origin of various citrus cultivars including lemons.
Materials and Methods
Plant Materials and Genomic DNA Extraction
The citrus samples used in this research were obtained from two public institutes in the Republic of Korea: Citrus Research Institute, National Institute of Horticultural and Herbal Science, Rural Development Administration, and Agricultural Research and Extension Services, Jeju Special Self-Governing Province. Leaf tissues were rinsed with running tap water and then stored at -70°C until use. Genomic DNA (gDNA) was purified from the leaf tissue using the Biomedic® Plant gDNA Extraction Kit (Biomedic Co., Ltd., Bucheon, Korea). The quantity and quality of the purified gDNA were determined by the DeNovix DS-11+ spectrophotometer (DeNovix, Wilmington, DE, USA) and agarose gel electrophoresis.
Mining of Polymorphic Plastid InDel loci and Primer Design
In order to develop polymorphic InDel markers in the organelle genomes of citrus, we compared the complete plastid genomes of four citrus species, C. limon (GenBank accession No. NC_034690), C. maxima (NC_034290), C. reticulata (NC_034671), and C. sinensis (NC_008334) by multiple sequence alignment using the CLC Genomics Workbench (ver. 6.8.4; Qiagen, Aarhus, Denmark). Putative polymorphic InDel markers were preliminarily screened by routine PCR using gDNAs from representative varieties of the four citrus species. The PCR reaction and cycling conditions were described in our previous report (Woo et al., 2019). PCR products were separated on a 2.5% (w/v) agarose gel to confirm PCR amplification and polymorphism among the varieties tested.
Genotyping by M13-tailed PCR and Data Analysis
The M13-tailed PCR method was used for genotype analysis using the selected polymorphic InDel markers (Schuelke, 2000). The PCR reaction condition was the same as in our previous report (Woo et al., 2019). The cycling conditions for PCR amplification followed the protocol described previously by Schuelke (2000). Fragment analysis of the PCR products was described previously (Kim et al., 2012). Calling of allele sizes was performed using the GeneMapper software (ver. 4.0; Applied Biosystems, Foster City, CA, USA). The unweighted pair group method with arithmetic mean (UPGMA) dendrogram was constructed using MEGA software (v. 7.0) (Kumar et al., 2016), which is embedded in PowerMarker (v.3.25; Liu and Muse, 2005). Genetic parameters such as major allele frequency, number of alleles, genetic diversity, and polymorphic information content were measured by calculating the shared allele frequencies using PowerMarker software (v. 3.25; Liu and Muse, 2005).
Results and Discussion
Development of Polymorphic InDel Markers from Citrus Plastid Genomes
Since the complete chloroplast genome of C. sinensis (L.) Osbeck ‘Ridge Pineapple’ was first reported in 2006 (Bausher et al., 2006), the complete plastid genomes of nine citrus species are currently available from the organelle genome resources of the National Center of Biotechnology Information (NCBI) (URL https://www.ncbi.nlm.nih.gov/ genome/browse#!/organelles/citrus). Recently, the complete chloroplast genome sequence of a medicinal landrace citrus ‘Jinkyool’ (C. sunki) in Jeju, Korea was also reported (Yoo et al., 2020). From the comparative in silico analysis of the whole-plastid genomes of C. limon, C. maxima, C. sinensis, and C. reticulata, we mined 20 putative major InDel loci (> 2 bp) specific to lemon or the other citrus species. From the primary screening by routine PCR using several citrus accessions of the four citrus species, we finally selected seven plastid InDel loci out of eight candidates (Table 1). Amplicon sizes of the selected InDel loci ranged from 252 (Limon#21) to 400 base pairs (bps) (Limon#16). All of the selected InDel loci were located in inter-genic spacers or introns (Table 1 and Suppl. Fig. 1s). This result corresponds to the fact that high-resolution of DNA barcodes for species identification and phylogenetic analysis in plants have been primarily found in the non-coding regions of the plastid genome (Hollingsworth et al., 2011; Jiao et al., 2019). The forward primers were tagged with 18-mer of the M13 sequence for efficient fragment analysis of the seven InDel loci by the cost-effective M13-tailed PCR method (Schuelke, 2000) (Suppl. Table 1s).
Table 1.
List of primers for the amplification of seven InDel loci
| Locus | Primer name | Primer sequence (5’ to 3’) | Expected amplicon size (bp) | Target regionz |
| Limon#01 | Limon#01-F2 | GCGCATACCAACAATATCAT | 261 |
310 to 570 (trnH-GUG/psbA) |
| Limon#01-R2 | TGCATGAACGTAATGCTCAT | |||
| Limon#13 | Limon#13-F1 | AGGGTCGGTCTTGAAACA | 325 |
34,198 to 34,522 (trnE-UUC/trnT-GGU) |
| Limon#13-R1 | AAGGCCAAAAAGCCCCTT | |||
| Limon#15 | Limon#15-F1 | CTAACAATTACGAGAATCTAG | 351 |
49,783 to 50,133 (trnT-UGU/ trnL-UAA) |
| Limon#15-R1 | CGAATTAGAATAGAGCAAATTT | |||
| Limon#16 | Limon#16-F2 | AGTGATATGGCTCGCCATA | 400 |
51,821 to 52,220 (trnF-GAA/ ndhJ) |
| Limon#16-R1 | ATGCCTGAAAGTTGGATAGG | |||
| Limon#21 | Limon#21-F1 | TATCGAGGGGCTTTTCTTC | 252 |
75,351 to 75,602 (2nd intron of clpP) |
| Limon#21-R1 | ATCAAAATCGGGCGAATCC | |||
| Limon#22 | Limon#22B-F1 | TAGTGTCCTTGCCCATGA | 255 |
84,574 to 84,828 (rps8/rpl14) |
| Limon#22B-R1 | AACTCGAGTTTTTGGTGC | |||
| Limon#23 | Limon#23-F2 | TGTAGACCCTCGCAATAGTT | 325 |
87,397 to 87,721 (rps3/rpl22) |
| Limon#23-R2 | ATGGGTCCTACTGCGAAA |
Discrimination of lemons from Other Citrus Groups using Polymorphic InDel Markers
In this study, we used 46 citrus accessions belonging to lemon, grapefruit, mandarin, pummelo, sour orange, orange, papeda, tangor, and tangelo groups, which were obtained from two public institutes in Korea (Table 2). Seven polymorphic InDel markers were applied to the citrus accessions to determine their genotypes (Suppl. Table 2s). Table 3 shows the genetic characteristics of the plastid InDel loci based on the genotype analysis of 46 citrus accessions. A total of 23 alleles, ranging from 2 (Limon#13, Limon#21, and Limon#22) to 5 (Limon#15 and Limon#23) per locus, were observed among the 46 accessions, with an average of 3.3 alleles per locus. Major allele frequency (MAF) varied from 0.37 to 0.76. The average genetic diversity (GD) value was 0.53, ranging from 0.36 to 0.74, and the average polymorphism information content (PIC) value was 0.47, ranging from 0.30 (Limon#13, Limon#21, and Limon#22) to 0.69 (Limon#23).
Table 2.
Citrus samples used in this study
Table 3.
Characteristics of the seven plastid InDel loci based on the genotype analysis of 46 citrus accessions including 17 lemon resources
| Locus | SS | NOBS | Availability | NG | MAF | NA | GD | PIC |
| Limon#01 | 46 | 46 | 1 | 4 | 0.59 | 4 | 0.58 | 0.52 |
| Limon#13 | 46 | 46 | 1 | 2 | 0.76 | 2 | 0.36 | 0.30 |
| Limon#15 | 46 | 46 | 1 | 5 | 0.37 | 5 | 0.73 | 0.68 |
| Limon#16 | 46 | 46 | 1 | 3 | 0.59 | 3 | 0.56 | 0.50 |
| Limon#21 | 46 | 46 | 1 | 2 | 0.76 | 2 | 0.36 | 0.30 |
| Limon#22 | 46 | 46 | 1 | 2 | 0.76 | 2 | 0.36 | 0.30 |
| Limon#23 | 46 | 46 | 1 | 5 | 0.37 | 5 | 0.74 | 0.69 |
| Mean | 46 | 46 | 1 | 3.3 | 0.60 | 3.3 | 0.53 | 0.47 |
SS, sample size; NOBS, number of observations; Availability is defined as 1-OBS/n, where OBS is the number of observations and n is the number of individuals sampled.; NG, Genotype number; MAF, major allele frequency; NA, number of alleles; GD, genetic diversity. GD, often referred to as expected heterozygosity, is defined as the probability that two randomly chosen alleles from the population are different.; PIC, polymorphism information content.
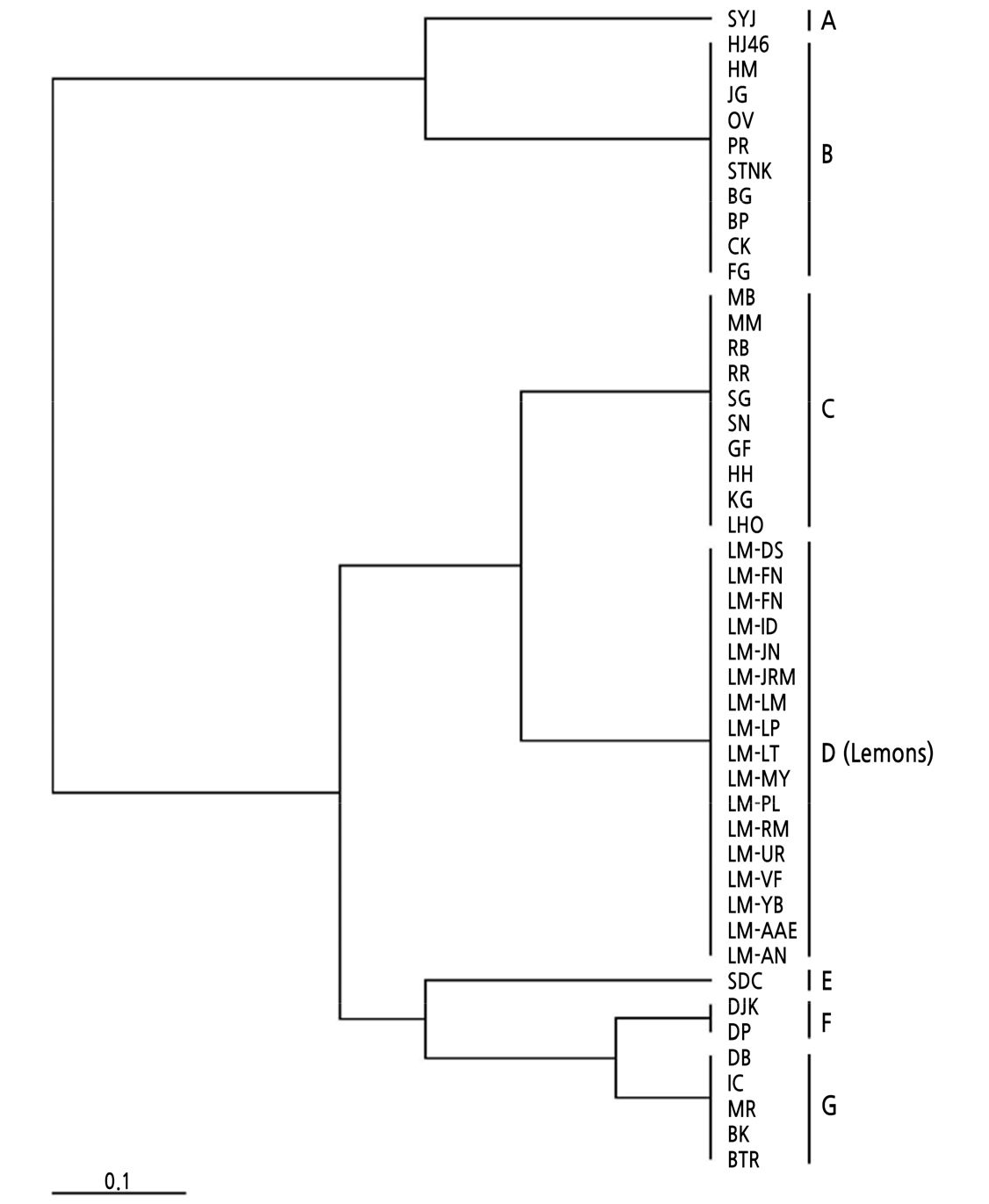
A total of 23 alleles derived from the seven polymorphic InDel loci were used to analyze the genetic relationships among the 46 citrus accessions. An UPGMA dendrogram was constructed based on the genetic similarity matrices among the accessions. Fig. 1 illustrates the results of the cluster analysis based on the genotype data of the seven plastid InDel loci. The resulting dendrogram revealed that the accessions used in the analysis could be distinctly classified into seven groups, forming a separate lemon cluster. This result suggests that these seven plastid InDel loci comprise a robust marker set to reliably discriminate scion cultivars from lemon rootstocks. Phylogenetic analysis using mitochondrial InDel and chloroplastic SSR markers showed that all C. limon accessions investigated clustered to the sour orange group with C. aurantium and C. limetta, etc., and were separated from the other five groups, which were the citrons (including C. medica only), C. micrantha (including C. aurantifolia and C. aurata, etc.), edible mandarins (including C. kinokuni, C. deliciosa, and C. clementina, etc.), wild mandarins (including C. sunki, C. reticulata, and C. limonia, etc.), and pummelos (including C. maxima and C. paradisi, etc.) (Curk et al., 2016). This report supports our dendrogram forming a lemon group that is separated from other citrus species (Fig. 1). Similar tree topology was also observed from the dendrogram of citrus genetic resources that were constructed using 17 nuclear SSR markers (Woo et al., 2019; Woo et al., 2020).
Molecular genetic evidences using intergeneric sexual hybrids revealed the maternal inheritance pattern of chloroplast DNA in citrus (Moreira et al., 2002; Abkenar et al., 2004). Our data also support a maternal pattern of organelle inheritance in citrus. As an example, cluster B contained three mandarins (BG, JG, and OV), five tangors (BP, CK, HM, PR, and HJ46), and two tangelos (STNK and FG). Tangors and tangelos originated from the genetic crosses of mandarin x C. sinensis and mandarin x C. maxima (or mandarin x C. paradisi), respectively (Park et al., 2020). Especially, three tangors (BP, HM, and PR) and one tangelo (STNK) shared Citrus hybrid ‘Kiyomi’ (CK) as a grandmother, which originated from the genetic combination of C. unshiu and C. sinensis (Park et al., 2020). According to the Swingle system, C. unshiu is considered to be a group of mandarin varieties (Froelicher et al., 2011). Our result suggests that all citrus cultivars of cluster B share mandarin as a female parent, further supporting the maternal inheritance of the chloroplast genome in Citrus.
DNA barcoding is an effective tool that enables rapid and accurate identification of plant species. However, single-locus DNA barcodes lack adequate variation in closely related taxa. Recent accumulation of whole-plastid genome sequences within closely related taxa has allowed researchers to develop cost-effective and reliable barcodes for accurate plant identification (Li et al., 2015). As shown in this study, reliable, multi-locus barcodes from the comparative analysis of whole-plastid genomes can increase our ability to distinguish closely related horticultural taxa such as citrus.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20210057_Figure_1s.pdf
Multiple sequence alignment from seven InDel target region from the plastid genomes of 4 citrus species, C. limon, C. maxima, C. sinensis and C. reticulata. Green arrows indicate forward and reverse primers amplifying InDel locus.
- HORT_20210057_Table_1s.pdf
List of primers for the genotype analysis of seven InDel loci by the M13-tailed PCR. Bold sequences indicate M13 tag
- HORT_20210057_Table_2s.pdf
Allele size data of 46 citrus accessions determined by seven polymorphic InDel markers