Introduction
Materials and Methods
Molecular modeling
Molecular modeling study
Construction of transient plant expression vectors for RVGP production
Transient expression in N. benthamiana leaves using the syringe agro-infiltration method
PCR analysis
Reverse transcription-PCR (RT-PCR)
SDS-PAGE and western blot analyses
Enzyme-linked immunosorbent assay (ELISA)
Results
Molecular modeling of the RVGP-FcK fragment
Confirmation of the presence of the RVGP-FcK gene in N. benthamiana plant leaves at different positions (top, middle, and base) and time points (1, 3, 5, 7, and 9 dpi)
Confirmation of RVGP-FcK gene transcription and protein expression in N. benthamiana leaves infiltrated with Agrobacterium
Transient expression of the RVGP-FcK protein in agro-infiltrated leaves (at the top, middle, and base leaf positions) at 1, 3, 5, 7, and 9 dpi
Comparison of the expression levels of the RVGP-FcK proteins in the total soluble proteins of agro-infiltrated leaves (top, middle, and base) at 1, 3, 5, 7, and 9 dpi
Confirmation of the assembled RVGP-FcK proteins isolated from agro-infiltrated leaves
Discussion
Introduction
The rabies virus (RABV), scientific name Rabies lyssavirus, a negative-stranded RNA-virus belonging to the genus Lyssavirus, family Rhabdoviridae, is primarily transmitted through the bites of rabid animals and can infect various mammals, including bats, skunks, raccoons, and foxes (Knobel et al., 2005; Mahamat et al., 2012; Velasco-Villa et al., 2017). Rabies induces neurological infections when it spreads through muscle and subcutaneous tissues (Wormser and Fisher, 2003). Effective treatment measures can prevent the onset of rabies (Martinez, 2000). RABV is a single negative-stranded RNA virus composed of five proteins, including a glycoprotein (G) (Tordo et al., 1986). The RABV glycoprotein (RVGP) plays a critical role in virus transmission and infection due to its transmembrane spike structure (Roche et al., 2008; Zhang et al., 2013). Thus, RVGP is a protein known for its immunogenic antigen sites and is considered to be a potential vaccine candidate (Wiktor et al., 1973) to stimulate the production of virus-neutralizing antibodies (Prehaud et al., 1988). Thus far, recombinant RVGP vaccines have been produced in cell systems (Ertl, 2009) and researched as a next-generation vaccine option (Starodubova et al., 2015). Results include the induction of a humoral immune response and the generation of neutralizing IgGs (Hu et al., 2006; Astray et al., 2014).
Plant expression systems are recognized as viable alternatives for producing immunotherapeutic proteins (Burnett and Burnett, 2020). These systems are more cost-effective compared to mammalian cell expression systems given that they require a less complex infrastructure. Plants primarily need water, soil, and light for growth. Once the plant expression system is established, maintaining plants under suitable environmental conditions is not prohibitively expensive (Ma et al., 2003). Furthermore, plant expression systems eliminate the risk of zoonotic pathogens (Shahid and Daniell, 2016; Lee et al., 2020), and plant-derived proteins tend to be more stable due to certain post-translational modifications compared to bacterial expression systems (Masson and Rossignol, 1995; Burnett and Burnett, 2020). Additionally, the stable mass production of plant cell lines is relatively straightforward (Ma et al., 2005; Lee and Ko, 2017). To express valuable recombinant proteins in plants using Agrobacterium tumefaciens, there are two main approaches: transient expression and transgenic plant expression (Marsian and Lomonossoff, 2016). Transient expression via agro-infiltration has is becoming more preferred in the manufacturing of biologics in plants due to the cost-effectiveness and rapid protein production capabilities associated with this method (Mardanova et al., 2017; Norkunas et al., 2018). Typically, transient plant expression systems yield a higher amount of protein per unit weight of plant tissue compared to transgenic plant expression systems (Kim et al., 2021; Lim et al., 2022; Kang et al., 2023; Oh et al., 2023). However, the level of recombinant protein expression is influenced by the leaf position and the time that elapses after infiltration, as demonstrated in previous research (Yamamoto et al., 2018; Kim et al., 2021).
In this study, we transiently expressed RVGP fused with the human immunoglobulin G Fc fragment, which was tagged with the endoplasmic reticulum (ER) retention signal KDEL (RVGP-FcK), in N. benthamiana plants. The purpose of this is to elucidate the spatial and temporal factors that optimize the production of RVGP-FcK, a promising vaccine candidate against the rabies virus.
Materials and Methods
Molecular modeling
The 3D structure of RVGP-FcK was predicted using the AlphaFold2 code (Fig. 1). The RVGP component used in this study had a topology similar to the previously resolved structure of the RVGP strain CVS-11 (PDB id: 6LGW). The engineered Fc region was connected by a long linker segment, suggesting that the two domains, i.e., RVGP and Fc, could exist in various configurations. The geometry of the bound antibody was modeled based on the structure of the RVGP complexed with a neutralizing antibody (PDB id: 6LGW). The structure of the bound antibody was visualized using the X-ray crystal structure of immunoglobulin G (PDB id: 1HZH). The dashed circle in Fig. 1B indicates the epitope region of RVGP and the variable domains (VH and VL) of the anti-rabies antibody bound to the epitope region. The molecular graphic figure was produced by PyMOL (http://pymol.org). RVGP (Gene ID: 1489856, NCBI) was cloned, and the gene was synthesized.
Molecular modeling study
Structural prediction of RVGP-FcK was undertaken using ColabFold v1.5.2 based on MMseqs2 and AlphaFold2, an AI-based protein structure prediction software package (Mirdita et al., 2022). The sequence of RVGP-FcK was input into the pipeline, employing the ‘pdb100’ template mode, which uses a database of PDB structures filtered at 100% sequence identity. We set the number of relaxations to 5 to refine the predicted model. For multiple sequence alignment (MSA), the ‘mmseqs2_uniref_env’ and ‘unpaired_paired’ modes were utilized. Both the RVGP component and the Fc region demonstrated high confidence levels with predicted IDDT (pIDDT) scores exceeding approximately 80. The predicted topology of RVGP closely resembled the known structure of the rabies virus glycoprotein (strain CVS-11, PDB id: 6LGW [ref: Cell Host Microbe 2020, 27, 441]). Segments of the linker region and disordered terminal loops which exhibited low confidence scores indicative of structural flexibility were omitted from the final model to enhance the clarity of Fig. 1. The potential binding geometry between RVGP-FcK and the antibody was modeled based on the RVGP structure complexed with a neutralizing antibody (PDB id: 6LGW [ref: Cell Host Microbe 2020, 27, 441]). This reference structure provided a template with which to understand the interaction interface and guide the complex generation process. The molecular graphic figure was generated using PyMOL v2.5.4.
Construction of transient plant expression vectors for RVGP production
RVGP cDNA (Gene ID: 1489856, NCBI) was synthesized for the antigen as a recombinant rabies virus vaccine candidate by Bioneer (Daejeon, South Korea) and fused with the human immunoglobulin G (IgG) Fc fragment along with the KDEL endoplasmic reticulum retention sequence (Figs. 1 and 2). This fusion, known as RVGP-FcK, was designed as a candidate for a recombinant rabies virus vaccine. RVGP was then amplified by polymerase chain reaction (PCR) with infusion primer forward: 5′- CAA ATT CGC GAC CGG ATG GCT ACT CAA CGA AGG GCA AAC -3′, reverse: 5′-TGC GGC CGC ACC GGT TAC ATA CTT CCC CCA GTT CGG GAG A -3′. The PCR primers used here were synthesized by Macrogen (Seoul, South Korea). The RVGP PCR products were fused to the gene encoding IgG FcK pEAQ vector by infusion (Takara Infusion Kit, Seoul, South Korea) to generate pEAQ RVGP-FcK. The RVGP-FcK gene was then inserted into the transient plant expression vector, pEAQ, resulting in the generation of the pEAQ RVGP-FcK vector. Within the pEAQ RVGP-FcK vector, the expression of RVGP-FcK was regulated by the cauliflower mosaic virus (CaMV) 35S promoter (35S-P), which included a duplicated enhancer region (E/35S-P) (as illustrated in Fig. 2). The gene-carrying vectors were subsequently transferred into Escherichia coli DH5α for cloning purposes.
Fig. 1.
Illustration of the 3D structure of RVGP-FcK and bound antibody. The predicted 3D structure of RVGP-FcK is depicted in a rainbow color scheme for the RVGP component and in gray for the engineered Fc region. The RVGP component is observed to have a topology similar to that of a previously resolved structure of the rabies virus glycoprotein (strain CVS-11, PDB id: 6LGW [ref: Cell Host Microbe 2020, 27, 441]). The engineered Fc region is connected via a long linker, indicating that the two domains, i.e., RVGP and Fc, could adopt various configurations. Dotted lines between the RVGP and the Fc region imply this flexible and long linker segment. The bound antibody is represented in a lighter shade to delineate its constant (CL, CH1, CH2, CH3) and variable (VL, VH) domains. The geometry of the antibody bound to RVGP was modeled using the structure of the RVGP complexed with a neutralizing antibody (PDB id: 6LGW [ref: Cell Host Microbe 2020, 27, 441]). The structure of the antibody was visualized by referring to the X-ray crystal structure of immunoglobulin G (PDB id: 1HZH [ref: Science 2001, 293, 1155]). The dashed circle highlights the epitope region on the RVGP and the variable domains of anti-rabies antibodies, which are critical for the specificity and affinity of the antibody-antigen interaction.
Transient expression in N. benthamiana leaves using the syringe agro-infiltration method
The transient expression vector pEAQ RVGP-FcK was introduced into A. tumefaciens (LBA4404) through electroporation (Fig. 2). Subsequently, 100 µL of A. tumefaciens was transferred into 35–40 mL of Luria–Bertani (LB) broth supplemented with 100 mg·L-1 of kanamycin and cultured for 24 hours at 28°C with continuous shaking at 200 rpm. Following cultivation, pre-cultured Agrobacterium (with an optical density reading of 0.3–0.4) was centrifuged at 3,000 rpm for 15 minutes. The resulting Agrobacterium cell pellet was then diluted in an infiltration buffer composed of 10 mM MES (pH 5.5), 10 mM MgCl2, and 0.1 mM acetosyringone (Oh et al., 2023). The cells, suspended in the infiltration buffer, were introduced into the leaves of 16-week-old N. benthamiana plants using a 1-mL needle syringe (Fig. 2). To achieve this, a small incision was gently made on the abaxial side of the leaf without puncturing it. Subsequently, the infiltration buffer was introduced into the leaves through this incision. To analyze the time-dependent and leaf-position-dependent recombinant protein expression levels, leaves from the top (#1, 2, and 3 leaves), middle (#4, 5, and 6 leaves), and bottom (#7 and 8 leaves) positions that had undergone infiltration were sampled at one, three, five, seven, and nine days post-infiltration (dpi) (Fig. 2). All harvested leaves were preserved at –80°C for subsequent analyses. The agro-infiltration experiments were repeated three times.
Fig. 2.
Schematic diagram of the transient plant expression systems used to produce the RVGP-FcK protein as a vaccine candidate, and the physical morphology of agro-infiltrated leaves (top, middle, and base) at 1, 3, 5, 7, and 9 dpi. Transient plant expression vectors used for producing RVGP-Fc with the ER retention signal KDEL (RVGP-FcK). RVGP-FcK gene expression cassette in the pEAQ vector: Cauliflower mosaic virus (CaMV) 35S promoter; SP, endoplasmic reticulum (ER) signal peptide; RVGP, rabies virus glycoprotein; IgG Fc, human immunoglobulin G Fc; K, ER retention motif, KDEL; NOS-T, nopaline synthase terminator. Agro-infiltration of Agrobacterium carrying the transient expression vector pEAQ RVGP-FcK into N. benthamiana leaves (top, middle, and base). Pre-cultured (O.D. of 0.3–0.4) transformed Agrobacterium in a needle. A small nick was made on the abaxial side of 16-week-old N. benthamiana leaves. The Agrobacterium solution carrying the pEAQ RVGP-FcK vector was infiltrated into the extracellular spaces of the leaf using a needle syringe. Physical morphology of agro-infiltrated leaves (top, middle, and base) as observed at 1, 3, 5, 7, and 9 dpi.
PCR analysis
Leaf tissue samples (100 mg) harvested from the infiltrated N. benthamiana plants were used to extract DNA using a DNA extraction kit (RBC Bioscience, Seoul, Korea). PCR amplification was performed to confirm the presence of the recombinant gene encoding RVGP-FcK. The following primer pairs were used (target, forward primer, reverse primer): the RVGP fragment in pEAQ RVGP-FcK (an infusion primer was used), 5′-CAA-ATT-CGC-GAC-CGG-ATG-GCT-ACT-CAA-CGA-AGG-GCA-AAC-3′, 5′-GTG-CAT-CCT-TCG-TCC-TCC-ACT-ACC-3′; the IgG-Fc fragment in pEAQ RVGP-FcK, 5′-CAG-GAC-TGG-CTG-AAT-GGC-AAG-3′, 5′-CCA-TTG-CTC-TCC-CAC-TCC-AC-3′; and the KDEL fragment in pEAQ RVGP-FcK, 5′-CAA-CTA-CAA-GAC-CAC-GCC-TCC-3′, 5′-CTC-ACC-AAA-CAT-AGA-AAT-GCA-CAC-3′. The PCR conditions for the positive control in E. coli DH5α were as follows: for pEAQ RVGP-FcK, initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing (at 61.5°C for the RVGP fragment of pEAQ RVGP-FcK, 59.9°C for the IgG Fc fragment, and 58.3°C for the KDEL fragment) for 10 s, and an extension at 72°C for 20 s. Then, a final extension step was performed at 72°C for 5 min. The non-infiltrated plant DNA samples and the pEAQ-RVGP-FcK vector were amplified using identical PCR sets as negative and positive controls, respectively.
Reverse transcription-PCR (RT-PCR)
The transcript of RVGP-FcK was identified in the leaves of the transient plant expression system using RT-PCR. Total mRNA was extracted from N. benthamiana leaf samples harvested using a QuantiTect Reverse Transcription kit (Qiagen) according to the manufacturer’s protocol. RT-PCR was conducted using a Maxime PCR Premix Kit (Intron Biotechnology, Seoul, Korea). Complementary DNA (cDNA) was PCR-amplified to confirm the mRNA transcription of the RVGP-FcK gene in the leaves of the transient plant expression systems. The following primer pairs were used (target, forward primer, reverse primer): the RVGP fragment in pEAQ RVGP-FcK, 5′-CAA-ATT-CGC-GAC-CGG-ATG-GCT-ACT-CAA-CGA-AGG-GCA-AAC-3′, 5′-GTG-CAT-CCT-TCG-TCC-TCC-ACT-ACC-3′; the RVGP fragment in pBI121 RVGP-FcK, 5′-GCT-CCA-CTA-ACC-ACG-ATT-ACA-CC-3′, 5′-GGG-AAG-AGG-AAG-ACT-GAC-G-3′; the IgG-Fc fragment in pEAQ RVGP-FcK and pBI121 RVGP-FcK, 5′-CAG-GAC-TGG-CTG-AAT-GGC-AAG-3′, 5′-CCA-TTG-CTC-TCC-CAC-TCC-AC-3′; the KDEL fragment in pEAQ RVGP-FcK, 5′-CAA-CTA-CAA-GAC-CAC-GCC-TCC-3′, 5′-CTC-ACC-AAA-CAT-AGA-AAT-GCA-CAC-3′; and the KDEL fragment in pBI121 RVGP-FcK, 5′-CAA-CTA-CAA-GAC-CAC-GCC-TCC-3′, 5′-GAG-TTC-ATC-TTT-ACC-CGG-GGA-CAG-3′. The RT-PCR conditions for pBI121 RVGP-FcK and pEAQ RVGP-FcK were as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing (at 61.5°C for the RVGP fragment of pEAQ RVGP-FcK, 59.9°C for the IgG Fc fragment, and 58.3°C for the KDEL fragment) for 10 s, and an extension at 72°C for 20 s. Then, a final extension step was performed at 72°C for 5 min. The RT-PCR conditions for the RVGP fragment in pBI121 RVGP-FcK were as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 10 s, and an extension at 72°C for 50 s, after which a final extension step was performed at 72°C for 5 min. Non-infiltrated leaves were used as negative controls. The elongation factor 1-α (EF-1α) gene was included as a reference gene.
SDS-PAGE and western blot analyses
The procedure for protein extraction from both the infiltrated (with Agrobacterium carrying pEAQ-RVGP-FcK) and non-infiltrated N. benthamiana leaves (130 mg) involved grinding each sample with 390 µL of 1X PBS in a 1.5-mL tube using a homogenizer to extract the total soluble proteins. Following homogenization, 96 µL of the homogenized sample was mixed with 24 µL of a 5X protein sample buffer (Shin et al., 2022). All samples were denatured at 122°C for 10 min. The total soluble proteins (TSP) were separated through SDS-PAGE and transferred to a nitrocellulose membrane (Millipore) (Song et al., 2021; Kang et al., 2023). For immunoblotting, the membrane was initially treated with 5% skim milk (Sigma) at room temperature (RT) for three hours. The blot was then incubated with rabbit anti-RVGP antibody (MyBioSource, San Diego, CA, USA) diluted at a 1:1,000 ratio in 5% skim milk for 90 minutes at RT. Following this incubation step, the blot was washed with a 1X TBS-T buffer three to four times, with each wash lasting 15 minutes. Subsequently, the blot was incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (H+L) antibody (Jackson Immuno Research) diluted at a 1:5,000 ratio in 5% skim milk and then incubated for 90 minutes at RT. The membrane was further incubated with HRP-conjugated goat anti-human IgG Fcγ fragment antibody (Jackson Immuno Research) diluted at a 1:5,000 ratio in 5% skim milk for 90 minutes at RT. The proteins in both blots were detected using a chemiluminescence substrate (BioRad), and the bands were visualized on X-ray film (Fuji, Tokyo, Japan).
Enzyme-linked immunosorbent assay (ELISA)
ELISA was conducted to compare the expression levels of the RVGP-FcK proteins by measuring the binding of RVGP and FcK to anti-RVGP and Fc antibodies, respectively. Leaves from the transient plant system were ground in 1X PBS using a homogenizer. MaxiSorp Clear Flat-Bottom Immuno Nonsterile 96-well plates (Nunc, Rochester, NY) were coated with anti-rabies RVGP antibodies (50 ng/well) in a coating buffer (0.1 M carbonate-bicarbonate buffer, pH 9.6). The plates were then incubated overnight at 4°C. After incubation, the plates were washed three times with 1X PBS-T (1X PBS and 0.05% v/v Tween 20; 200 µL/well) and blocked with 3% BSA diluted in 1X PBS-T for two hours at 28°C. Leaf extracts from the transient plant systems (100 µL/well) were serially diluted in a blocking buffer (dilution factors: 1/80, 1/40, 1/20, 1/10, 1/5, and 1) and incubated for two hours at room temperature (RT). Non-infiltrated leaf extracts served as negative controls. Each plate was treated with HRP-conjugated rabbit anti-human IgG Fc antibody (1:5,000) to bind the Fc region of RVGP-FcK in the total soluble proteins and incubated for two hours at 37°C. Next, 100 µL/well of TMB peroxidase substrate (KPL, Gaithersburg, MD) was added to the plate in each case and incubated for 45 seconds. The reaction was stopped by adding 100 µL/well of TMB stop solution (Seracare, Milford, CT). Finally, the absorbance at 450 nm was measured using an Epoch microplate spectrophotometer (Biotek). To investigate whether the plant-derived RVGP-FcK (RVGP-FcKP) proteins were intact to interact with both anti-rabies virus mAb and the anti-human IgG Fc antibody, purified RVGP-FcKP, E. coli-derived RVGP (RVGPE), and human-derived IgG-Fc (IgG-FcH) were included in the assay. This ELISA allowed us to assess the binding of RVGP-FcK to both anti-RVGP mAb and anti-Fc mAb in the transient plant expression system, providing valuable insights into its expression levels and interactions. The ELISA experiments were repeated three times.
Results
Molecular modeling of the RVGP-FcK fragment
We conducted molecular modeling studies to understand the structural aspects of the RVGP-FcK fragment sequences using anti-RVGP mAb (Fig. 1A and 1B). As depicted in Fig. 1B, RVGP-FcK forms one side of the IgG molecule. It is divided into two main regions: the RVGP region, which contains the antibody binding sites, and the Fc region, which is responsible for various effector functions. The structure of the RVGP-FcK has yet to be fully elucidated. However, given the high sequence identity in the Fc region, we constructed a model based on the well-characterized structure of the human immunoglobulin G (IgG) b12 (PDB id: 1HZH). The formation of this complex was modeled using X-ray crystallographic data from the complex structure of RVGP and anti-rabies virus mAb. We circled the RVGP bound to the anti-rabies virus mAb SO57 to mark its position and significance during the interaction (Fig. 1B).
Confirmation of the presence of the RVGP-FcK gene in N. benthamiana plant leaves at different positions (top, middle, and base) and time points (1, 3, 5, 7, and 9 dpi)
The N. benthamiana leaves were harvested according to their positions (top, middle, and base) and time points (1, 3, 5, 7 and 9 dpi) (Figs. 2 and 3). In general, young, mature, and old leaves were positioned at the top, middle, and base of the plant, respectively (Figs. 2 and 3). The presence of the RVGP-FcK gene in the agro-infiltrated leaves was confirmed by means of a PCR analysis (Fig. 3B). Specific primers were used to detect different parts of the RVGP, human IgG Fc, and KDEL of the RVGP-FcK gene. All agro-infiltrated leaves, regardless of their position and time point, exhibited the expected amplified bands of RVGP, Fc IgG Fc, and FcK (KDEL) (Fig. 3). No bands were detected in non-agro-infiltrated leaves.
Fig. 3.
PCR analysis to confirm the presence of the RVGP-FcK gene in the transient plants based on the number of days post-infiltration (dpi) and the position of the leaf (top, middle, and base). (A), (B), and (C) PCR analysis to confirm the presence of the RVGP gene in pEAQ (197 bp), IgG-Fc (224 bp), IgG-FcK (KDEL) (269 bp) in the top, middle, and base of the plant, respectively. A total of 2 μL of each sample was loaded. +, positive control, pEAQ RVGP-FcK in E. coli DH5α; -, negative control, non-transient N. benthamiana plant (NT); Day 1–9, RVGP-FcK from the transient plant leaf based on the number of days post-infiltration (dpi).
Confirmation of RVGP-FcK gene transcription and protein expression in N. benthamiana leaves infiltrated with Agrobacterium
To verify the mRNA and protein expression of the RVGP-FcK gene in N. benthamiana leaves infiltrated with Agrobacterium carrying pEAQ RVGP-FcK (Suppl. Fig. S1A), we conducted a RT-PCR analysis. Specific PCR primers were employed to detect the RVGP, human IgG Fc, and RVGP-FcK genes. The transient plant expression system produced the expected sizes for the amplified RVGP (197 bp), IgG Fc (224 bp), and IgG FcK (KDEL) in pEAQ (269 bp) (Suppl. Fig. S1A). RT-PCR products with the anticipated sizes for both RVGP and IgG Fc were detected in the leaves of the transient plant expression systems. Notably, the agro-infiltrated leaves exhibited band signals for both RVGP and IgG Fc around five and seven days post-infiltration (dpi). No transgenes were detected in the non-agro-infiltrated leaf. The EF-1α band, utilized as a standard control, displayed the expected size. In order to confirm protein expression in the agro-infiltrated leaves, we conducted a western blot analysis (Suppl. Fig. S1B). This analysis revealed the presence of primarily two protein bands at approximately 80 kDa. Notably, at 5 dpi, the band around 80 kDa exhibited the highest density among all time points (Suppl. Fig. S1B). This observation confirmed the expression of the RVGP-FcK protein in the agro-infiltrated leaves.
Transient expression of the RVGP-FcK protein in agro-infiltrated leaves (at the top, middle, and base leaf positions) at 1, 3, 5, 7, and 9 dpi
Western blotting was also used to determine the protein expression levels of RVGP-FcK (~80 kDa) in the total soluble proteins extracted from the agro-infiltrated leaves depending on the leaf position and dpi (Fig. 4). The leaves expressed RVGP-FcK (~80 kDa), RVGP (~55 kDa), and FcK (~25 kDa). The RVGP protein, used as a positive control, was detected at ~60 kDa using the rabbit anti-rabies glycoprotein antibody (Fig. 4, left panels) but was not detected using the anti-human IgG-Fc antibody (Fig. 4, right panels). In the anti-Fc IgG treatment, the ~80 kDa protein band was consistently detected at 5, 7, and 9 dpi in the top leaves (Fig. 4, top, left panel). In the agro-infiltrated middle leaf, the density of the ~80 kDa band density was strongest at 5 and 7 dpi and was weaker at 9 dpi (Fig. 4, middle, left panel). No ~ 80 kDa band was detected in the base leaves at any time point (Fig. 4, base, left panel). In the anti-RVGP IgG treatment, the density of the ~80 kDa protein band was strongest at 7 dpi. Nevertheless, the protein band was evenly detected at 5, 7, and 9 dpi in the top leaves (Fig. 4, top, right panel). However, in the middle leaf, the density of the ~80 kDa band was strongest at 5 dpi (Fig. 4, base, right panel), and it decreased with an increase in the dpi (Fig. 4, middle, right panel). No ~ 80 kDa band was detected in the base leaves at any dpi (Fig. 4, base, right panel). Overall, the relative protein expression was consistently high in the top leaf position. However, the strongest band density was observed in the middle leaves at 5 dpi (Fig. 4B).
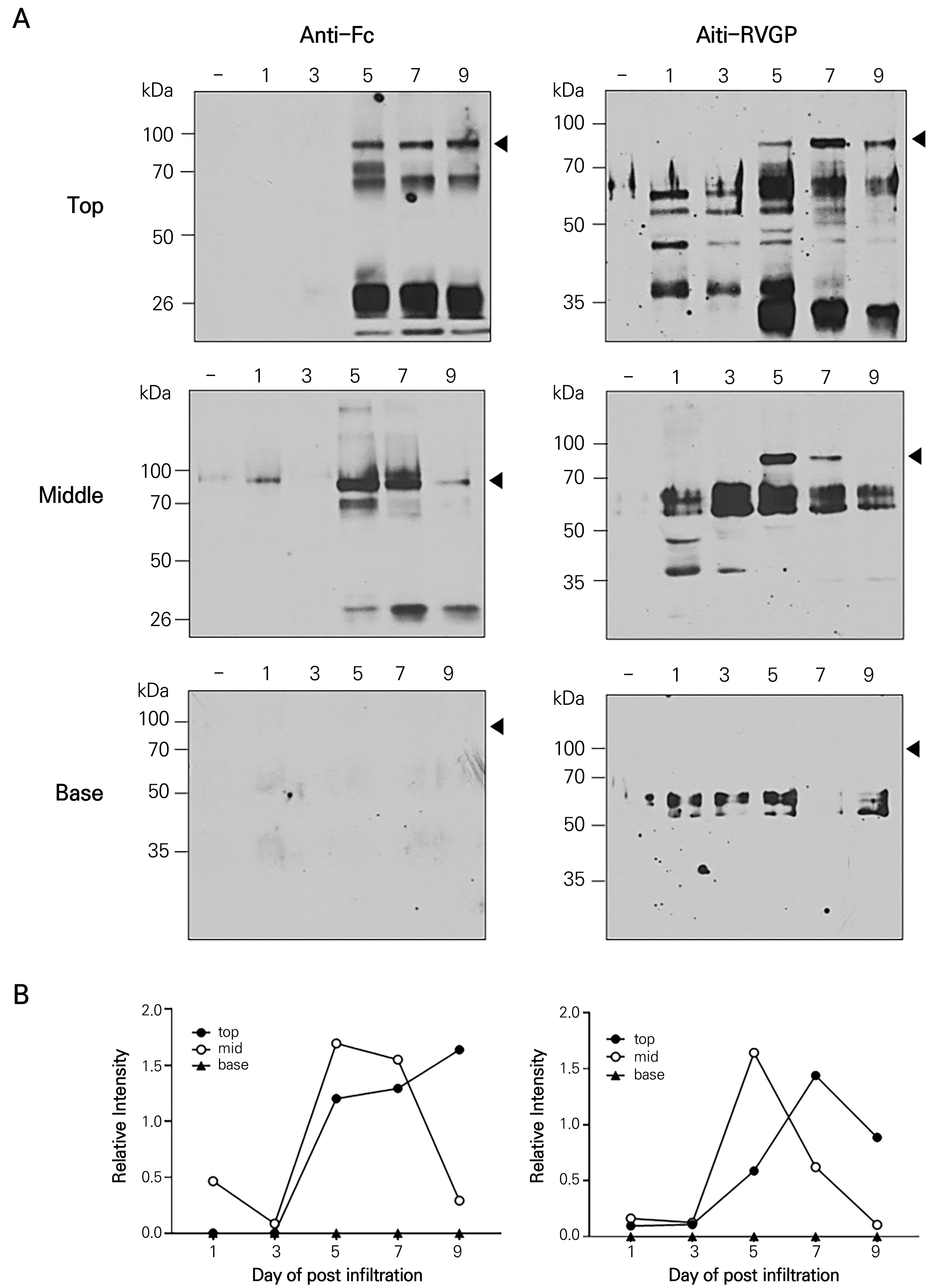
Fig. 4.
Immunoblot analysis of RVGP-FcK expressed in transient plant leaves (top, middle, and base) at 1, 3, 5, 7, and 9 dpi. The total soluble proteins (TSP) were separated using an 8% SDS-PAGE gel. +, positive control (RVGPE) (A) and (EpCAM-FcH) (B); -, non-agro-infiltrated leaf; 1, 3, 5, 7, and 9, agro-infiltrated leaves at 1, 3, 5, 7, and 9 dpi. An immunoblot analysis was conducted to confirm the expression levels of RVGP-FcK (~80 kDa).
Comparison of the expression levels of the RVGP-FcK proteins in the total soluble proteins of agro-infiltrated leaves (top, middle, and base) at 1, 3, 5, 7, and 9 dpi
The absorbance signal was not detected in any leaf positions until 1 dpi (Fig. 5). At 3 dpi, the absorbance signal was detected in both the top and middle leaves at 3 dpi (Fig. 5). At 5 and 7 dpi, the strongest absorbance signals were observed in the top leaves (Fig. 5). At 9 dpi, relatively high absorbance signals were still observed in both the top and middle leaves (Fig. 5). In the base leaves, no absorbance signal was observed until 5 dpi, while a trace signal was detected at 7 and at 9 dpi.
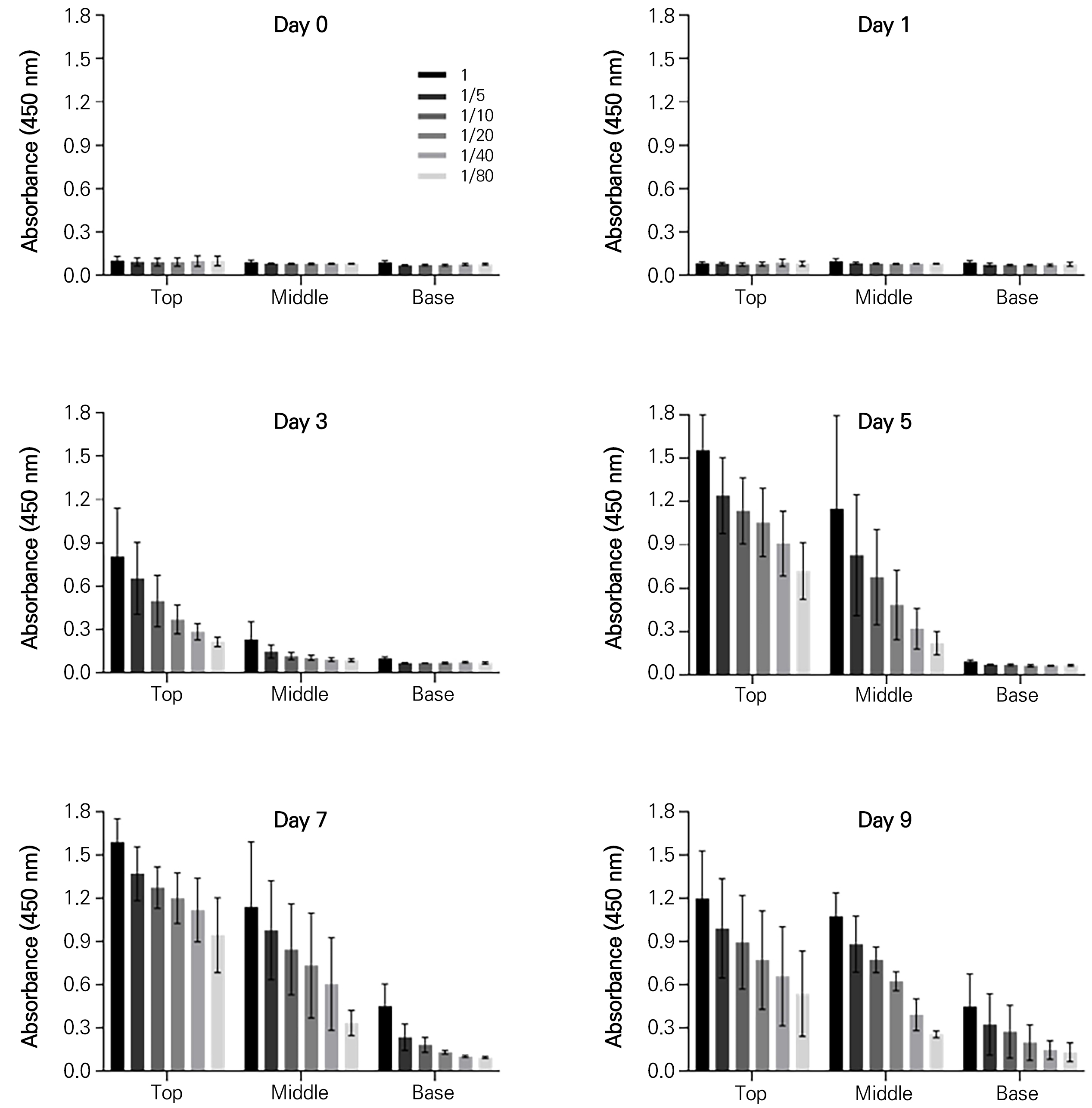
Fig. 5.
ELISA to compare the expression levels of the RVGP-FcK protein at different leaf positions (top, middle, and base) and at various time points (1, 3, 5, 7, and 9 dpi). NT, non-transient plant leaf extraction sample; Day 1–9, transient plant leaf extraction samples at 1–9 dpi; top, middle, and base leaf positions of N. benthamiana plant. The expression levels of RVGP-FcK for the top, middle, and base positions were assessed using ELISA. A statistical analysis was conducted and significance is indicated by * (p < 0.05) and ** (p < 0.01).
Confirmation of the assembled RVGP-FcK proteins isolated from agro-infiltrated leaves
A sandwich ELISA analysis was conducted to confirm the assembly of RVGP-FcK proteins isolated from agro-infiltrated leaves using an anti-rabies glycoprotein antibody and an HRP-conjugated anti-IgG Fc antibody (Fig. 6). The assembly was confirmed through the binding of anti-RVGP IgG to RVGP and the binding of Fc antibodies to FcK (Fig. 6A). A 96-well plate was coated with anti-RVGP antibody (50 ng/well) and E. coli-derived RVGP (RVGPE), human IgG-Fc (IgG-FcH), and RVGP-FcK proteins isolated from agro-infiltrated leaves (RVGP-FcKP) were applied to the wells (Fig. 6A). In the RVGP-FcKP protein treatment, the absorbance signals were observed relative to the dilution factor (Fig. 6B). However, in the RVGPE and IgG-FcH cases, no absorbance signals were observed (Fig. 6B).
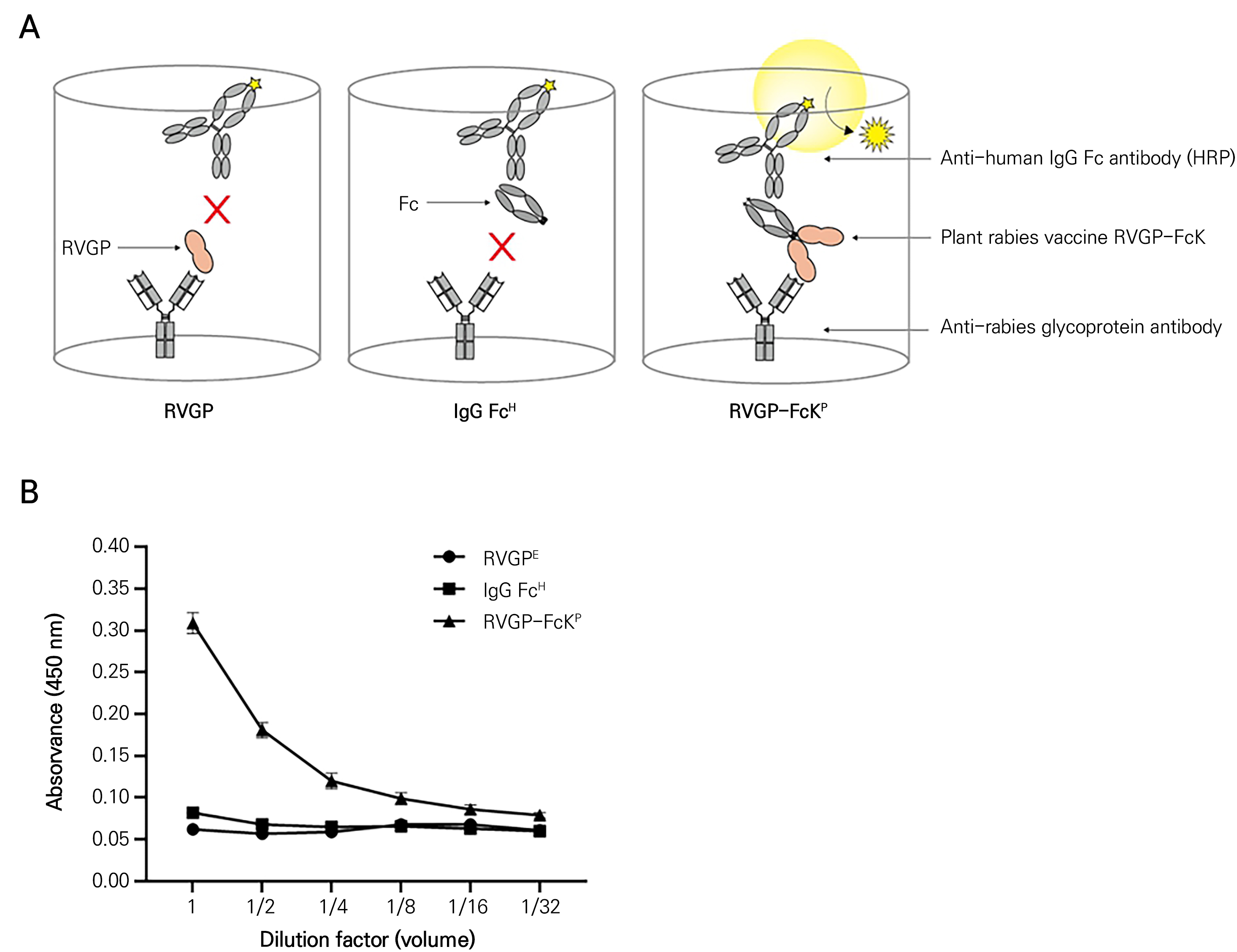
Fig. 6.
ELISA conducted to confirm the binding activities of the RVGP-FcK protein to the anti-RVGP antibody and the anti-human IgG Fc antibody: (A) schematic diagram showing the binding of RVGP-FcK, RVGP, and human IgG Fc to the anti-RVGP-FcK and anti-human IgG Fc antibody conjugated with HRP. (B) ELISA was conducted to compare the concentration-dependent binding activities of RVGP-FcK. The titers were serially diluted two-fold. Triangular line, plant-derived RVGP-FcK; rectangular line, commercial human-derived IgG-Fc fragment; round line, commercial E. coli-derived RVGP. A statistical analysis was conducted and significance is indicated by ** (p < 0.01).
Discussion
This study demonstrated that the expression of the recombinant RVGP-FcK fusion protein, a vaccine candidate for the rabies virus, is affected by the position of the leaf tissue and the number of days post-infiltration in transient N. benthamiana plants. In a previous study, it was found that the expression of the colorectal cancer antigenic Fc fusion protein GA733-Fc varies spatially and temporarily (Kim et al., 2021). Therefore, to enhance the efficiency of producing valuable recombinant RVGP-FcK protein in the transient plant expression system, it is necessary to optimize the timing and location of leaf harvesting. First, we observed the morphological changes in the leaves. The base leaves at 5–7 dpi showed slight chlorosis and wilting, which affected the expression level of the recombinant protein (Kim et al., 2021). A PCR analysis confirmed the presence of the recombinant RVGP-FcK gene in the agro-infiltrated leaves, regardless of the leaf position or dpi. According to RT-PCR results, however, the transcription level of the RVGP-FcK transgene was detected at 3 dpi and reached its highest level at 5 dpi. A previous study by the authors found that the highest expression of the recombinant GA733-FcK gene occurs in N. benthamiana at 5 dpi (Kim et al., 2021). Thus, in this study, leaf samples were harvested at 5 dpi to confirm the mRNA expression of the RVGP-FcK gene. However, it is important to note that mRNA and protein levels are not always tightly correlated (Song et al., 2021). Thus, a western blot analysis was also conducted to determine the expression of the RVGP-FcK protein in relation to the leaf position and dpi in the transient plant expression system. The results showed that protein expression levels were significantly affected by the leaf position and dpi. In the top leaves, high expression of RVGP-FcK was observed at 5, 7, and 9, whereas in the middle leaves, low expression was observed at 9 dpi. Furthermore, an ELISA analysis was conducted to compare the expression levels of RVGP-FcK in relation to the leaf position and dpi more precisely. The ELISA analysis demonstrated that the expression of the RVGP-FcK protein was high in both the top and middle leaf positions. These results are consistent with those of previous studies involving citrus (Acanda et al., 2021) and avocado (Persea americana Mill.) (Salazar-González et al., 2023), which showed that transient protein expression levels are dependent on the leaf age and dpi. These results suggest that the top leaves at 5 and 7 dpi can be recommended for achieving the highest expression of the RVGP-FcK protein.
To confirm the Fc fusion form of the RVGP-FcK proteins isolated from agro-infiltrated leaves, a sandwich ELISA analysis was conducted with three different treatments: RVGPE, IgG FcH, and RVGP-FcKP. Only RVGP-FcKP showed an absorbance signal, whereas RVGPE and IgG FcH did not. These results were expected given that RVGPE and IgG FcH do not have the IgG Fc domain and RVGP, respectively. In addition, these results suggest that the RVGP-FcK protein expressed in plant leaves has the human IgG Fc fusion formation. The Fc fusion strategy has been widely used in the production of valuable recombinant proteins. The Fc domain promotes proper protein folding for protein A-based purification, enhances the half-life in the bloodstream, and increases the immunogenicity of the fused antigen by enhancing its binding to antigen-presenting cells (Martyn et al., 2009; Wieland and Ahmed, 2019). Accordingly, Fc-fused protein vaccines against MERS, SARS-CoV, SARS-CoV-2, H5N1 influenza, colorectal cancer, and prostate cancer have been developed. These vaccines with Fc fusion are more immunogenic than those without it (Du et al., 2007; Li et al., 2013; Sun et al., 2021).
In general, the expression of recombinant proteins is directly and indirectly affected by transcription, translation, and post-translational events such as folding, assembly, glycosylation, and targeting (Ma et al., 2003). In addition, the production of valuable recombinant proteins depends on the expression system used. Plant expression systems can be broadly categorized as plant nuclear (transgenic expression) and transient expression systems. Both systems offer advantages such as low-cost plant biomass production and no risk of human pathogen contamination. The transgenic plant system allows for easy scalability of plant biomass in that it uses a transgenic seed bank with stable transgene expression (Ko, 2014). In contrast, the transient plant system has limited potential for scaling-up. Moreover, the process of infiltrating Agrobacterium into plant leaf biomass is labor-intensive due to the transient expression of the recombinant protein gene in the plant cell cytosol, without stable insertion into the plant genomic DNA (Nosaki et al., 2021). Nevertheless, the transient plant system, when optimized with respect to the harvested leaf position and dpi, offers a range of superior advantages, such as high yields and rapid expression outcomes compared to the transgenic plant system (Nosaki et al., 2021).
In the present study, the Fc fusion protein RVGP-FcK was investigated as a recombinant vaccine candidate against the rabies virus. It was found that this protein could be transiently expressed and assembled in the N. benthamiana plant, and the optimal leaf position and dpi were determined for achieving high expression levels of the RVGP-FcK protein. Overall, the transient N. benthamiana plant expression system shows promise as a potential alternative for producing valuable therapeutic proteins, including vaccines.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20240023_Fig_S1.pdf
Supplementary Fig. S1. Results of RT-PCR and western blot analyses confirming the mRNA and protein expression of the RVGP-FcK gene in the transient plants: (A) confirmation of the presence of the RVGP gene (197 bp) and IgG-Fc (224 bp) fragment in the transient plant expression systems. A total of 2 µL of each sample was loaded. -1, negative control (non-infiltrated leaf); 1, 3, 5 and 7, infiltrated leaf samples at 1, 3, 5 and 7 dpi, respectively. A RT-PCR analysis confirmed the transcription of the transgene in the transgenic tobacco plant and the transient expression system at 5 dpi. Total mRNA was isolated from the leaves of the transient and transgenic plant expression systems. The EF-1α gene, a housekeeping gene, served as the reference gene. (B) The expression of the RVGP-FcK protein was observed in the infiltrated leaf samples at 1, 3, 5 and 7 dpi through a western blot analysis.


