Introduction
Materials and Methods
Plant Materials
Screening of Proteins Interacting with the AMS Gene
Screening of Proteins Interacting with CmZR1
Glutathione S-transferase (GST) Pull-Down Verification of the Y2H Assay
RNA Extraction and Transcriptome Sequencing
qRT-PCR
Bioinformatics Analysis of Melon CmPMEI Gene Family
Determination of PME Enzyme Activity
Results
Protein Interaction with the AMS Gene
Screening of Proteins Interacting with CmZR1 and GST Pull-Down Verification
Transcriptome Sequencing of Differentially Expressed Genes (DEGs)
Phylogenetic Analysis of Melon, CmPMEI
Prediction of Conserved Sequences and Gene Structure Analysis
Analysis of Pectin Methylesterase Inhibitor Genes
PME Enzyme Activity
Discussion
Conclusion
Introduction
Melon (Cucumis melo. L) is an important crop of the family Cucurbitaceae and is planted worldwide. Melon has high heterosis for fruit and seed production. Male sterility (MS) is one of the significant traits and has been reported in a wide range of higher plants. This plays a crucial role in low-cost hybrid seed production by eliminating the need for emasculation and cross-pollination. MS enhances heterosis in the plant breeding industry and forms an important area of study in developmental biology (Kaul, 1988; Sorensen et al., 2003). The occurrence of MS is closely related to pollen development. The formation of tapetum is the key to anther development. A number of principal tapetum transcription factors in anthers have been identified, including Dysfunctional Tapetum1 (DYT1) (Zhang et al., 2006; Feng et al., 2012), ABORTED MICROSPORES (AMS) (Sorensen et al., 2003; Xu et al., 2010; Xu et al., 2014), basic Helix-Loop-Helix (bHLH) transcription factors, Male Sterility 1 (MS1) (Wilson et al., 2001; Yang et al., 2007) in Arabidopsis, the ATP synthase subunit alpha (ATPA) gene in ramie (Boehmeria nivea) (Duan et al., 2009), the ATPA gene in non-heading Chinese cabbage (Jiang et al., 2019), the Zmms1 gene in maize (Lu et al., 2018), and the MS2 gene in wolfberry (Cheng et al., 2018). Mutation of these genes at different stages of development results in pollen abortion (Ma, 2005). Moreover, the abnormal expression of other genes, including RAFTIN1 and Aps (Zhai et al., 2018) in the anthers causes MS by affecting the early cell and tapetum formation, meiosis, or pollen maturation (Sanders et al., 1999; Wilson et al., 2001; Boavida et al., 2005; Hord et al., 2006; Zhang et al., 2007; Zhu et al., 2008; Wilson and Zhang, 2009).
AMS has been demonstrated to be a major regulator of pollen wall development. The pollen wall not only provides mechanical protection for male gametophytes but also protects them from microbial attacks and environmental stress; thus, playing a vital role in specific pollen stigma recognition (Zinkl et al., 1999; Scott et al., 2004). It has been reported that the AMS mutants of Arabidopsisthaliana failed to accumulate lipid spore powder protein precursors (Xu et al., 2014) and altered the expression of anther genes (Xu et al., 2010). AMS is highly expressed in the tapetum, pollen mitosis I, and two-cell pollen periods to regulate the secretion of the dull layer, the formation of pollen wall materials, and transport of the tapetum to the ventricle in microspores and immature pollen grains. Its absence results in the vacuolation of tapetum cells and degradation of microspores (Sorensen et al., 2003; Xu et al., 2010). Ma et al. (2012) compared the AMS anther transcriptome with those of Sporocyteless (SPL)/Nozzle (NZZ) and Excess Microsporocytes 1 (EMS1)/Extra Sporogenous Cells (EXS) anthers, revealing both overlapping and different regulatory gene sets, including transcription factors and other proteins. Using yeast two-hybrid screening, Sorensen et al. (2003) studied the interaction between ASH1-related 3 (ASHR3) with the putative MycbHLH transcription factor AMS, which is a key regulator of anther development and stamen length. In addition to AMS, zinc ribbon (ZR) protein (zinc finger protein transcription regulation family) and pectin methylesterase inhibitor (PMEI) protein play significant roles in pollen wall development (Borg et al., 2014; Jin, 2017). The C2H2-type zinc finger proteins involved in pollen development act on the mechanism regulating tapetum degradation (Vizcay-Barrena and Wilson, 2006). The gene encoding the C2H2 zinc finger protein, DZA1/DZA2 participates in the second meiosis process of spermatogenesis (Borg et al., 2014). Pectin is one of the main components of the plant cell wall (Carpita and Gibeaut, 1993). The wall of the pollen tube tip is composed of a single layer of pectin and does not contain cellulose or callose like other plant cell walls (Li et al., 1994; Ferguson et al., 1998). Pollen tube growth inhibition is associated with pectin esterification (Li et al., 1996). This leads to the subsequent degradation of pectin under the combined action of polygalacturonase and pectin lyase (Louvet et al., 2006; Pelloux et al., 2007). The early activity of the pectin methylesterase (PME) promoter in Brassica napus during the tetrad isolation indicated that the PME isoform might play a role in tetrad division (Albani et al., 1991). Mutations in Arabidopsis thaliana pollen-specific pectin methylesterase 1 (AtPPME1), a type I PME, resulted in the loss of a regular pollen tube tip shape and slower growth (Tian et al., 2006). The diversity of PME expressed in Arabidopsis pollen suggests that other members of this protein family are associated with the growth of pollen tubes and may influence different aspects of this process (Pina et al., 2005). Similarly, mutation of the type II PME gene, VANGUARD 1 (vgd1), which has the highest expression level in the Arabidopsis pollen tube, leads to a slow growth of the style and conduction pathway, leading to a decrease in male fertility (Jiang et al., 2005).
In melon, five single recessive genes, designated MS-1 to MS-5, were originally identified as responsible for Genic Male Sterility (GMS) (Bohn and Whitaker, 1949; McCreight, 1983; Lecouviour et al., 1990; Pitrat, 2002; Park et al., 2004). Sheng et al. (2017) used Specific Length Amplified Fragment Sequencing (SLAF-seq) to map AMS as the candidate gene for MS-5. These results suggest that AMS may be a key candidate gene for MS in melon. However, the molecular mechanisms involved in AMS regulation of MS in melon are still unclear. The objective of the present study, therefore, was to understand the molecular mechanisms of the AMS regulatory network. We systematically elucidated the AMS-mediated pollen abortion mechanisms using yeast hybridization to identify proteins with potential key roles in the regulation of microsporogenesis. The expression and interactions of these candidate genes were further investigated at different developmental stages of MS and MF (Male Fertility) flower buds using transcriptomics, qRT-PCR, and bioinformatics. The findings on the MS network regulatory system will provide crucial information for the study of MS study at the transcription level in melon.
Materials and Methods
Plant Materials
The MS and MF melon lines were provided by Dr. James D. McCreight, Department of Agriculture Crop Improvement and Protection Research Center in Salinas, CA, USA. These lines were planted in a plastic greenhouse at Bayi Agricultural University Experimental station (125.03° latitude and 46.58° longitude), Daqing, Heilongjiang Province, China. Use conventional water and fertilizer management for cultivation. Samples (10 g) of flower buds (the tetrad stage and mononuclear pollen stage) with diameters of 1 and 2 mm were collected from the plants for construction of the yeast hybridization cDNA library. The samples were frozen in liquid nitrogen at -80°C before transcriptome sequencing, qRT-PCR and PME enzyme activity determination. To reduce potential differences between samples, three samples were used for transcriptome sequencing analysis and qRT- PCR.
Screening of Proteins Interacting with the AMS Gene
The cDNA library was constructed by Matchmaker library and a screening kit (Clontech, Mountain View, CA, USA) was used according to the manufacturer’s instructions. Yeast one-hybrid (Y1H) experiments were performed as previously described (Dai et al., 2020). Briefly, the cDNA was cloned into the vector, pGADT7 to construct an AD-fusion library for the yeast strain Y1HGold, a strain that does not grow in the absence of uracil. The sequence of the AMS promoter was then cloned upstream of the reporter gene promoter. Subsequently, the bait vector, pAbAi containing the AMS core sequence was transformed into the Y1HGold strain. The cDNA pool with flanking end sequences homologous to the prey vector, pGADT7-Rec was constructed using SMART™ MMLV RT (Takara Bio, Japan). The cDNA and the linear pGADT7-Rec vector were then co-transformed into the Y1HGold strain. Finally, after co-transformation, the cells were plated on the SD/-Leu/AbA to select colonies based on the prey proteins which activated the AbAr reporter. Positive colonies were analyzed by colony PCR and sequencing.
Screening of Proteins Interacting with CmZR1
Yeast two-hybrid (Y2H) interactions were conducted using the mating yeast strain, Y187 carrying the full-length pGBKT7-CmZR1 bait construct with the AD fusion library at 30°C. The culture was plated on selective media Quadruple Dropout Supplements (QDO) containing X-a-Gal (20 mg·L-1) for the nutritional reporter genes, HIS3 and MEL1 to identify positive two-hybrid interactions. The putative positive clones were transformed with empty vectors, pGADT7 and pGADT7-Rec as control to verify the interactions. Y187 cells transformed with pGBKT7-CmZR1 full-length and deletion constructs were tested for autoactivation of the HIS3 and MEL1 reporter genes. The yeast double hybrid test refers to the method of Thorstensen et al. (2008).
Glutathione S-transferase (GST) Pull-Down Verification of the Y2H Assay
The PGEX-6P-1 plasmid was transformed into the E. coli BL21 strain. Isopropyl β-d-1-thiogalactopyranoside (0.6 mM) was added to the cells which were then incubated at 20°C for 4 h. The bacterial proteins were sonicated and purified by GST agarose beads and separated using SDS-PAGE to determine the expression of GST and the GST-CmPMEI1 fusion protein. The GST and CmPMEI1-GST proteins were detected by pull-down assay using 20 µL of samples from the control group or experimental group, 50 ng of GST protein or CmPMEI1-GST protein, 10 µL of CmZR1 solution, 1:50,000 of GST antibody, and exposure for 10 s. For the detection of the HIS-CmZR1 protein, 20 µL of sample from the experimental group, 50 ng of GST protein or CmPMEI1-GST protein, 10 µL of CmZR1 solution, 1:1,000 of HIS antibody, and hypersensitive exposure for 2 min. The proteins were transferred to a PVDF membrane for western blot analysis. The conditions for the western blot were as follows: 50 ng of CmPME-GST and GST protein, 1:5,000 dilution of GST antibody, exposure for 10 s, 50 ng of HIS-CmZR1 and HIS control protein, 1:1,000 dilution of HIS antibody, and hypersensitive exposure for 15 min.
RNA Extraction and Transcriptome Sequencing
Total RNA from the melon flower buds was extracted using an RNA Mini Kit (Tiangen, Beijing, China), according to the manufacturer’s protocol. The flower buds from MS and MF plants were mixed as a single biological replication, respectively. The experiments were performed in triplicate independently for the MS and MF lines.
mRNA was isolated from the total RNA using oligo-dT magnetic beads and fragmented with fragmentation buffer for the synthesis of cDNA. The short fragments were purified for the end repair, and then connected with sequencing adapters. Finally, the cDNA (-250 bp )in length were used for the PCR amplification. The sequences were quantified and determined for quality using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA)and ABI Step One Plus RT-PCR System (Thermo Fisher, Waltham, MA, USA)after which the cDNA libraries were sequenced using the Illumina sequencing platform HiSeq 4,000 (Beijing Biomarker, Beijing, China).
qRT-PCR
The qRT-PCR assay was performed to verify the gene screened from the yeast system. The total RNA for the transcriptome sequencing was used for the qRT-PCR also. Reverse transcription and PCR assays were performed using a Thunderbird SYBR qPCR Mix kit (TOYOBO, Osaka, Japan), according to the manufacturer’s instructions. The qRT-PCR reaction program is: pre-denaturation 95°C 60 s; 40 cycles (95°C, 15 s, 58°C, 30 s, 72°C, 60 s); dissolution curve (65°C, 5 s, 95°C, 5 s). The primer used in the qRT-PCR was shown in Table 1 and the assay was performed with three biological samples.
Table 1.
Primers of selected genes used in this study
Bioinformatics Analysis of Melon CmPMEI Gene Family
Sequence Retrieval and Analysis of the PMEI Gene Family
The CmPMEI gene was obtained from yeast two-hybrid results. Screening of thePMEI gene family was performed using the Cucurbitaceae database (http://cucurbitgenomics.org/organism/18) for Melon. Next, the reported amino acid sequences of PMEI proteins from Cucumis sativus L. and Arabidopsis thaliana (L.) Heynh. were retrieved using the information available on the National Center for Biotechnology Information (NCBI) protein database (https://www.ncbi. nlm.nih.gov/protein) and the UniProt protein database (https://www.uniprot.org/).
Sequence Alignment and Phylogenetic Tree Construction
Multiple sequence alignments of the amino acid sequences were performed using MEGA 7.0 software. The phylogenetic trees were constructed separately for the PMEI using the neighbor-joining method with the bootstrap values set at 1,000 replicates.
Prediction of Conserved Sequences and Gene Structure Analysis
The conserved domains in the sequences were predicted using the SMART database and their distribution was analyzed by IBS software version 2.0. The conserved motifs of PMEI were analyzed through the MEME (http://meme-suite.org/).
Characteristics of Gene Structure Analysis
The accession numbers, length of the coding sequence, and the amino acid numbers of PMEI were obtained from the SMART database (http://smart.embl-heidelberg.de/). The physical and chemical properties, such as molecular formula, molecular weight, and isoelectric point (pI) were obtained from ExPASy (http://web.expasy.org/cgi-bin/protparam/ protparam).
Determination of PME Enzyme Activity
The PME enzyme activity was determined as previously described (Yang et al., 2012). Briefly, pre-chilled 8.8% NaCl (5mL) was added to the samples (3 g) which were then homogenized in an ice bath. The sample was then centrifuged at 10,000 rpm, at 4°C for 2 min. The pH of the crude enzyme extract was adjusted to 7.5 with 0.1 M NaOH and the sample was stored store at 4°C for later use. To determine the activity, 0.5% (w/v) citrus pectin solution (4 mL) and 0.01% bromothymol blue (0.3 mL) were added to 300 µL of the crude enzyme extract. The color of the indicator was changed due to the action of the enzyme and the pectin. The absorbance at 620 nm was measured after 2 min of reaction, and the change in absorbance (∆A620/min·g) was used to indicate the enzyme activity. The experiments were performed in three biological replicates.
Results
Protein Interaction with the AMS Gene
The positive PCR products of the co-transformation separated on agarose gel 1.5% showed bands of 500–2,000 bp for the genes in the positive clones of the prey plasmid. In total, 15 positive clones interacting with the AMS core elements were identified by sequencing analysis (Dai et al., 2020). The sequences were investigated using BLAST (www.ncbi. nlm.nih.gove/BLAST) to identify potential homologs and protein functions. Among these 15 proteins, three were found to be unknown proteins with no functional annotations. However, sequence comparison showed that there was high homology of the LOC103496635 (MELO3C020021P1) gene identified by the Y1H system to zinc ribbon protein motif genes. Therefore, we hypothesized that LOC103496635 is the gene responsible for early pollen development and was named as C. melo L. Zinc Ribbon protein 1 (CmZR1).
Screening of Proteins Interacting with CmZR1 and GST Pull-Down Verification
The CmZR1 gene was amplified by PCR and electrophoresed to recover the whole sequence of the bait gene (Fig. 1). The recombinant plasmid-positive strain was cultured in combination with the DNA library, and the microscopic examination showed normal growth of the yeast cells (Fig. 1E). The conjugated growth and various sizes of the insertion fragments of the library were found by using the pressure screening and the general primer PCR system. The gene size carried by the aggregates in the conjugated growths was different, with a range of 500–2,000 bp (Fig. 1B–D).
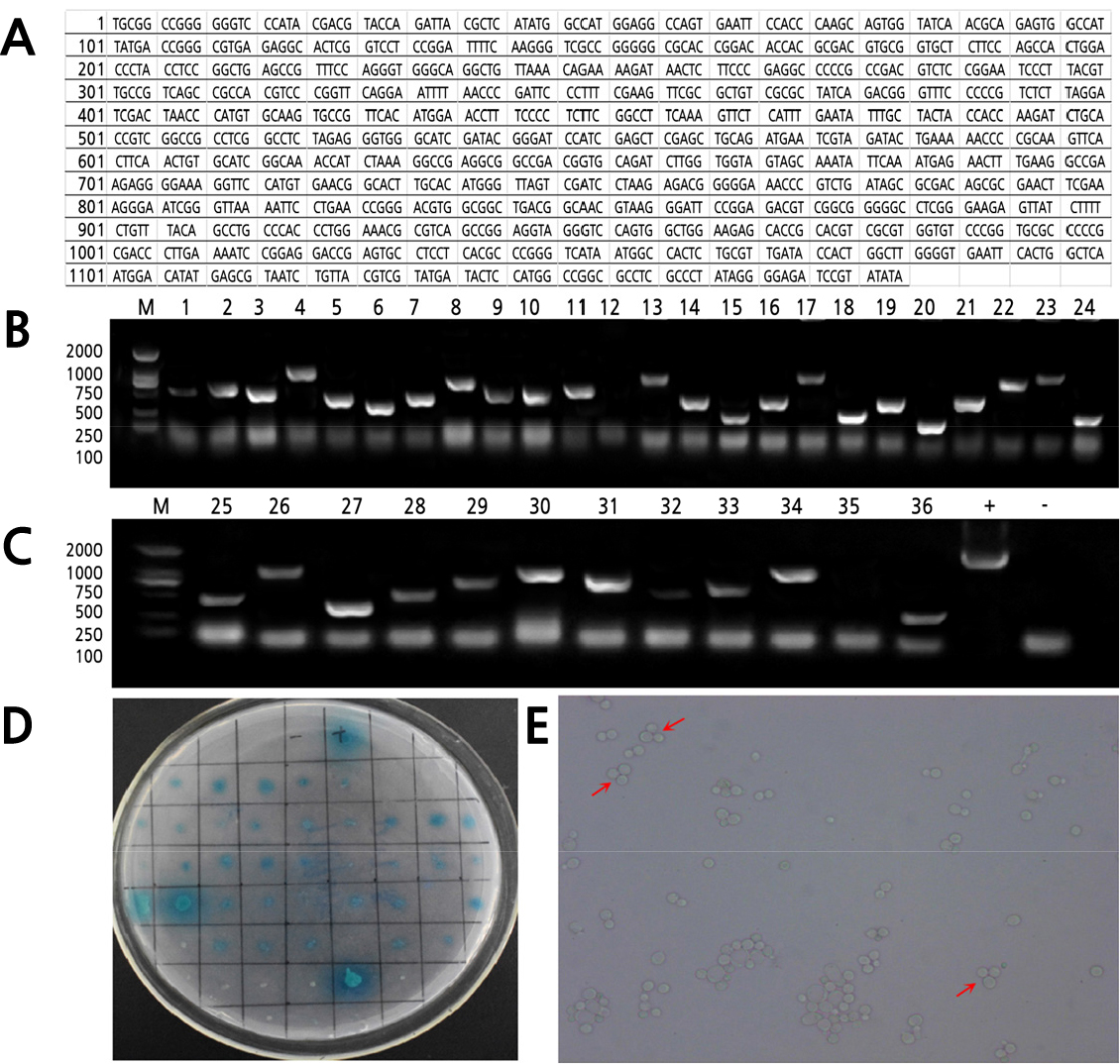
Fig. 1.
CmZR1 gene sequencing data and Y2H binding growth positive screening: CmZR1 gene sequencing results (A); Agarose gel electrophoresis maps of PCR products of positive colonies interacting with CmZR1 gene (B-C), M (Marker 2000/1000/750/500/250/100 bp), + (positive control), - (negative control); The combined growth yeast cells were transferred to QDO/X/A (SD/-Leu/-Trp/-His/-Ade/Aba/X) medium (D); Combined with growth yeast cell inverted microscope photographs (E).
A total of 19 proteins interacting with CmZR1 were identified by the Y2H system (some proteins were duplicated) (Table 2). The domains of these proteins were analyzed, showing that the 22 proteins had a high probability of having transcription factor activity. The aligned PMEI protein sequence closely resembled the LOC103483991 (MELO3C006821P1) gene sequence, named as CmPMEI1.
Table 2.
Functional annotations of positive clones for the Y2H system
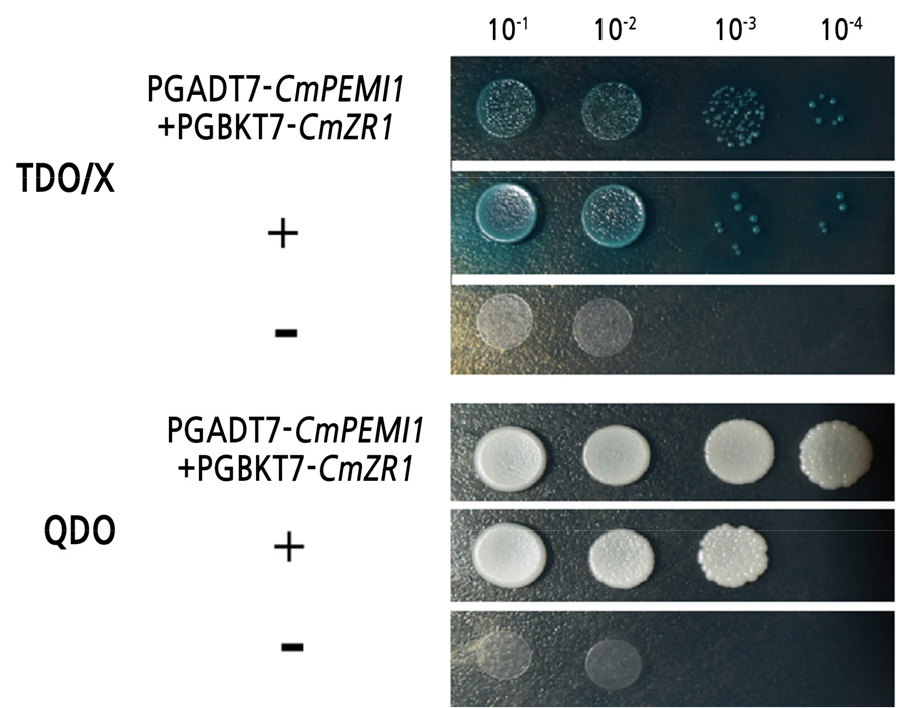
The point-to-point verification method was used to confirm further the reliability of the CmPMEI1 genes screened from the Y2H system. The bait plasmid pGBKT7-CmZR1 and the prey plasmid pGADT7-CmPMEI1 were co-transformed to Y2HGold yeast cells for the co-transformation and dot-plate verification. The recombinant bait plasmid showed no toxic effects on yeast cells and no self-activation effects on the Y2HGold yeast reporter gene suggesting an interaction between the CmPMEI1 and CmZR1 protein (Fig. 2).
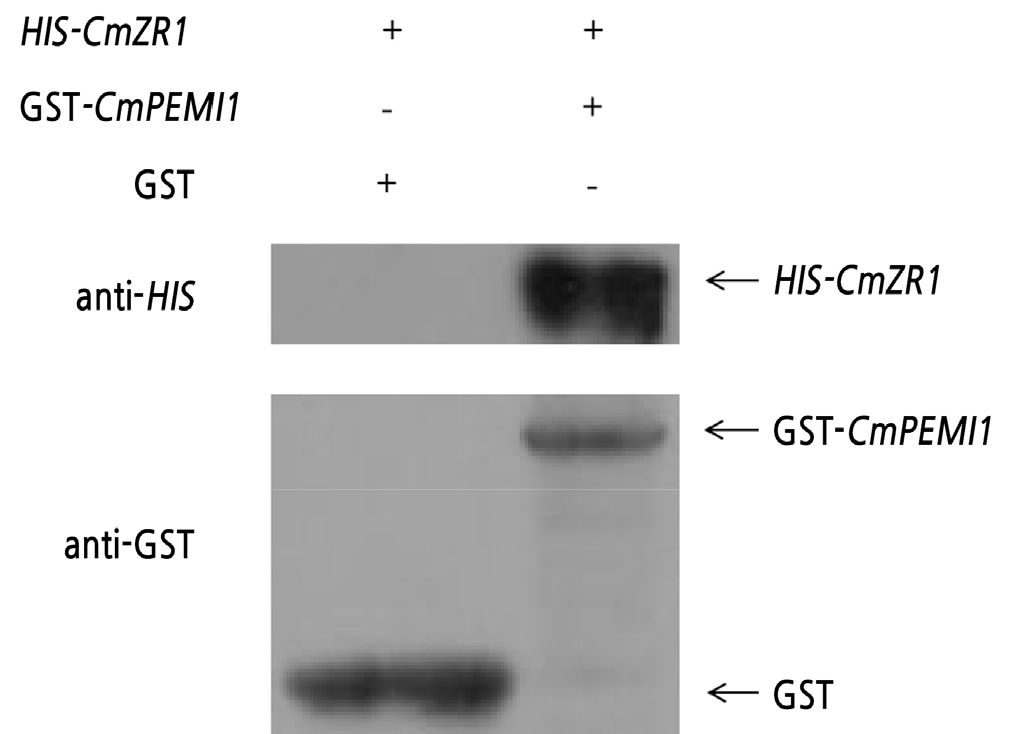
The GST pull-down test result is shown in Fig. 3. It can be concluded that GST-CmPMEI1 protein can interact with HIS-CmZR1 protein.
Transcriptome Sequencing of Differentially Expressed Genes (DEGs)
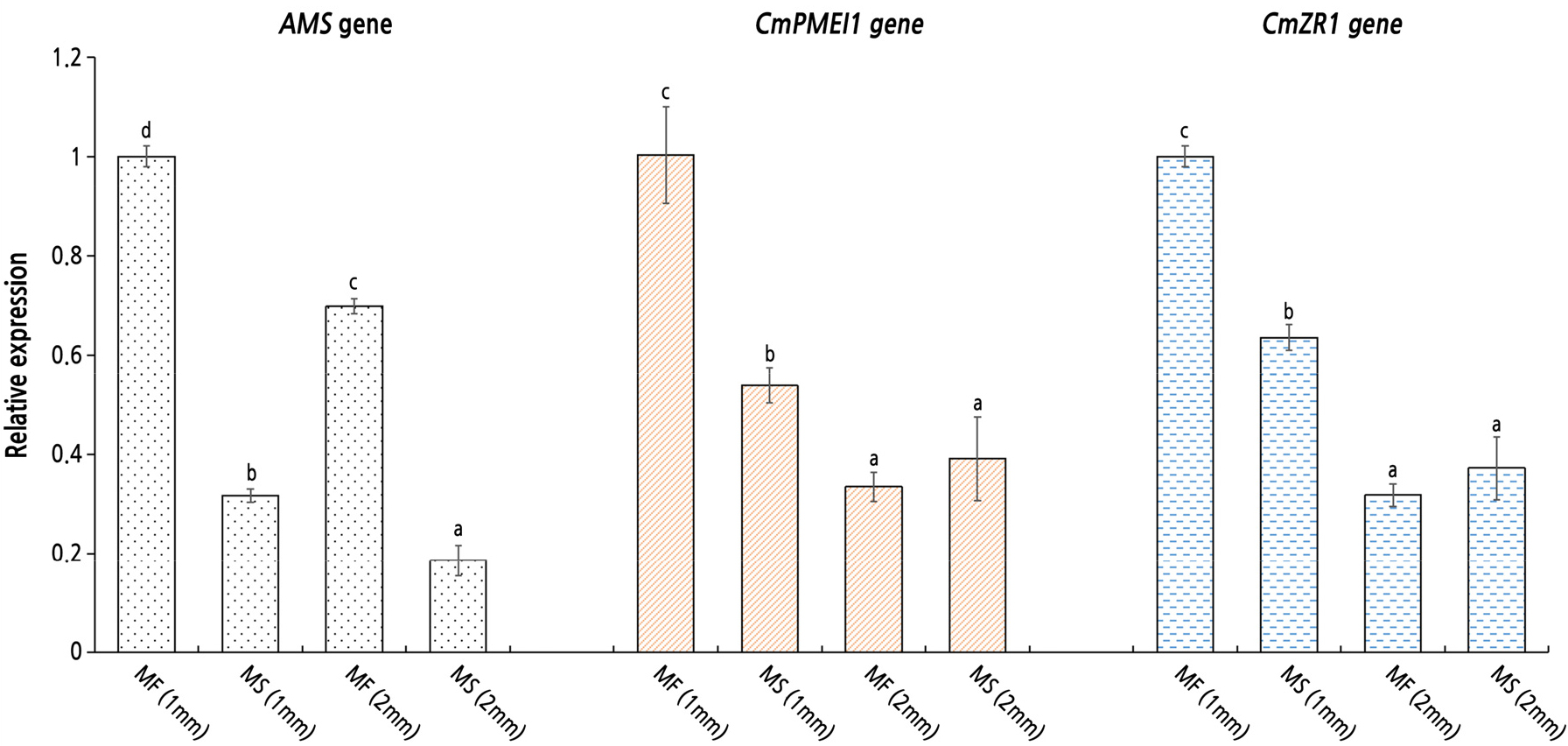
Twelve DEG libraries were constructed from the tetrad and monocyte stages. We detected a total of 820 differentially expressed genes (Dai et al., 2019). The genes, CmZR1 and CmPMEI1, were screened by the yeast system and subjected to qRT-PCR analysis. The qRT-PCR results showed that the expression of these genes was consistent with the sequencing results (Fig. 4). Compared with the MS plants, AMS was expressed significantly higher in the MF lines, whereas the CmZR1 gene was more highly expressed at the early stage of anther development in the MF lines. The expression trend of the CmPMEI1 gene was similar to that of the CmZR1 gene, with no expression difference between the MS and MF plants at the mononuclear pollen stage (Fig. 4).
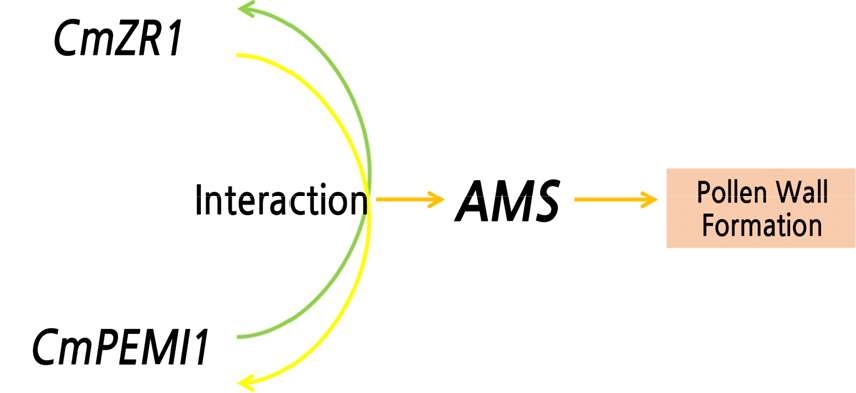
Although transcriptome sequencing revealed similar expression trends of the CmZR1 gene and the CmPMEI1 gene in MS and MF plants (Fig. 4), compared with the MF plants, the CmZR1 gene and the CmPMEI1 gene were expressed to a lesser extent in the MS plants at the early stage of mononuclear pollen development (length of flower bud = 2 mm), suggesting inhibition of CmZR1 and CmPMEI1 gene expression. The CmZR1 gene identified by the Y2H was located upstream of the AMS gene and interacted with the CmPMEI1 gene.
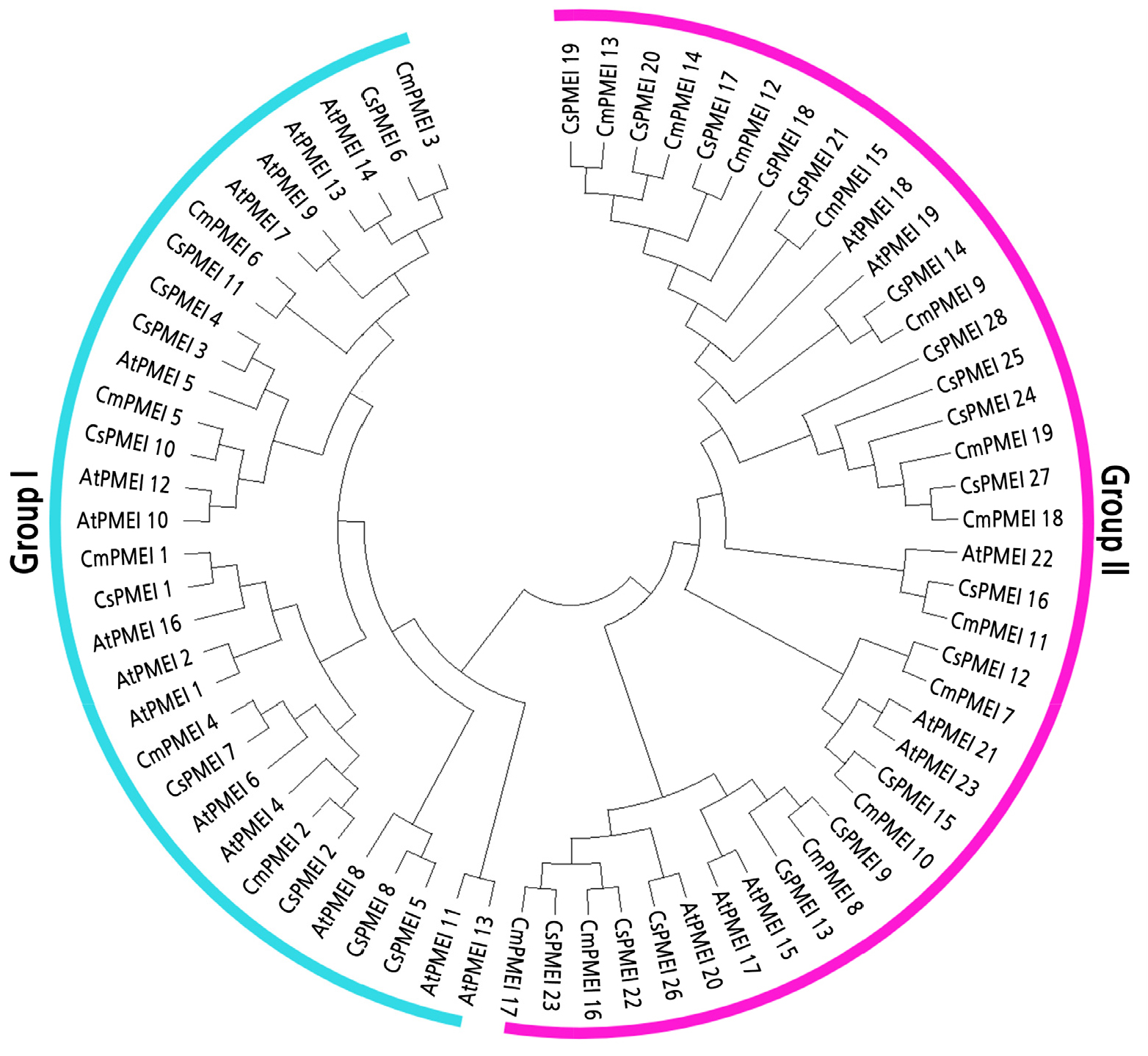
Phylogenetic Analysis of Melon, CmPMEI
CmPMEIs were divided into two sub-groups according to the phylogenetic tree (Suppl. Table 1s) (Fig. 6). The sub-group I contained 31 genes, including the CmPMEI1 gene screened by Y2H. The sub-group II consisted of 39 genes, mainly composed of PMEI genes of melon and cucumber. Within each sub-group, most melon PMEI and their cucumber homologs formed a phylogenetic branch at higher values.
Prediction of Conserved Sequences and Gene Structure Analysis
The conservative domain of melon PMEI genes analyzed by SMART software showed that the conserved domains of the melon PMEI genes were relatively simple, and had only one conserved domain PMEI (Suppl. Fig. 1s).
Analysis of Pectin Methylesterase Inhibitor Genes
The lengths of the PMEI genes ranged from 160 to 252 residues, with an average molecular weight of about 20,887 Da, and isoelectric points of 4.40-9.73. Subcellular localization analysis showed that most of the PMEI genes were expressed in chloroplasts and vacuoles (Suppl. Table 2s).
PME Enzyme Activity
For MF plants, the PME activity of tetrad stage (0.136 units) and mononuclear pollen stage (0.236 units) flower buds was significantly different (p < 0.05). For MS plants there was significant different of PME activity between tetrad stage (0.151 units) and mononuclear pollen stage (0.202 units) flower buds (p < 0.05). The PME enzyme activity of MS plants was higher than that of MF plants at the tetrad stage (length of flower bud = 1 mm). The PME enzyme activity of MF plants was higher than that of MS plants at the mononuclear pollen stage (length of flower bud = 2 mm). At the mononuclear pollen stage, the PME activity in MS plants was 14.7% lower than that of MF plants (Supp. Fig. 2s).
Discussion
Melon has a high intraspecific genetic variation and a small genome (450 Mb). Therefore, it has become a representative crop for genetic studies on important traits in the Cucurbitaceae family. Understanding the regulatory mechanisms of the MS network would be extremely beneficial for the utilization of heterosis. Several MS regulatory networks have been developed (Sorensen et al., 2003; Xu et al., 2010; Xu et al., 2014), in which the transcription factor AMS has been identified as a key candidate gene for MS (Ma et al., 2012; Xu et al., 2014; Xiong et al., 2016). AMS regulates different genes in the tapetum by forming complexes with other proteins such as DYT1-bHLH homodimers (Ma et al., 2012; Xu et al., 2014), thus affecting the normal expression of the transcription factor MS188.MS188 is a petal cell-specific transcription factor for pollen wall formation and interacts with AMS to form a complex for activation of the expression of CYP703A2 and other genes, leading to MS (Xiong et al., 2016). To the best of our knowledge, this is the first report, demonstrating a partial pathway related to the MS network in melon. Using a yeast expression system, we identified a number of proteins such as CmZR1 with potential key roles in the regulation of anther development and microsporogenesis. The expression and interactions of these candidate genes were further investigated at different developmental stages of MS and MF flower buds using transcriptome sequencing and qRT-PCR. The CmZR1 gene upstream of the AMS gene interacted with the CmPMEI1 gene leading to the male sterility in melons (Fig. 5). These findings in melon provide the information necessary to study the MS network at the transcription level in melon.
The CmZR1 gene belongs to the zinc finger protein family and participates in zinc ribbon protein synthesis regulation. Zinc finger proteins self-fold to form a “finger”-like structure that binds to RNA/DNA molecules, thus playing a vital role in the plant development and response to environmental stresses (Chen et al., 2019; Li et al., 2020). Previous studies have revealed the involvement of zinc finger proteins such as C2H2-zfp and BcMF20 in the development of the tapetum, sperm cell differentiation, and meiosis (Kapoor et al., 2002; Kapoor and Takatsuji, 2006; Borg et al., 2014; Han et al., 2018). Besides, Denes et al. (2000) demonstrated a preferential expression pattern of pollen development-related factors C2H2-zfp (Maz1 gene) in the early stage of Arabidopsis anther development. The mutation in the Maz1 leads to the abnormal deposition of the cytoplasm in the tetrad stage has established an essential role in the defective formation of the cytoplasm and microspores, leading to the reduction of pollen wall formation and pollen fertility (Lyu et al., 2019). The specific zinc finger transcription factor, the inhibition of BCMF20 expression in Arabidopsis mutants with BCMF20 gene deletion resulted in impaired pollen germination as well as low germination and seed setting rates (Han et al., 2018). Here, we studied the expression of the zinc finger protein gene CmZR1 upstream of AMS in the early stage of melon flower bud development. The CmZR1 gene was located upstream of AMS and affected the regular expression pattern, ultimately leading to MS in melon.Transcription factors bind not only to protein promoter regions but also to other proteins, to form a complex. In the present study, the results suggest that the interaction between the CmPMEI1 and CmZR1 genes upstream of the AMS gene regulates abnormal expression of the AMS gene, resulting in MS in melon (Fig. 5).
Observation of different flower buds development stages of melon indicated that the male flower differentiation was in the tetrad stage when the diameter of melon bud was less than 1.5 mm, in the mononuclear pollen stage when the diameter of melon bud was less than 2 mm, and pollen mature stage when the flower bud was ≈ 4mm (Wang et al., 2009). Previous research indicated significant different were detected in tetrads periods in pollen development for male sterility and fertility lines, because of pollen sac were empty in male sterile plants and pollen grain were not export may reduce male sterility in melon ms5 (Sheng et al., 2016). PME is a key enzyme in pectin metabolism (Louvet et al., 2006; Pelloux et al., 2007) and regulates the plant cell wall’s mechanical strength and chemical properties through demethylation of pectin, thus promoting cell development. The cell wall consists of a polymeric network of crystalline cellulose microfibers embedded in the hydrophilic matrix of hemicellulose and pectin (Denes et al., 2000). Previous studies have demonstrated that PME has a key role in the modification of the cell wall and regulates various physiological processes (Kagan-Zur et al., 1995; Guglielmino, 1997; Wakeley et al., 1998; Futamura et al., 2000; Micheli et al., 2000; Ren and Kermode, 2000; Li et al., 2002). Furthermore, studies by Paynel et al. (2014) showed that Kiwi PMEI inhibits PME activity in Arabidopsis and regulates the root elongation and simultaneously induces the pollen tube burst (Paynel et al., 2014). The change in mechanical strength and stiffness of the pollen tube wall are also considered to impact the elongation of pollen tube and its interaction with female flower tissues (Franklin-Tong, 1999). The PME enzyme activity of MS plants was higher than that of MF plants at the tetrad stage, but for mononuclear pollen stage, the PME enzyme activity of MS plants was lower than that of MF plants .Although the significant increasing of PME activity were all appeared in both MS and MF plant when comparing tetrad and mononuclear pollen stages, respectively, but there were no significant different between 2mm stage in MS vs MF plants. For investigated if PME activity were related to male sterility in pollen development stages, PME activity was also detected in pollen mature stage (flower bud was ≈ 4mm) and significant difference between MS and MF plants in this stage were identified (Suppl. Fig. 2s). based on the results, the PME enzyme may related to pollen development which induce male sterility of melon. And the similar results were also identified in Chinese Cabbage- pak-choi (Liu, 2006) .
By analyzing the bioinformatics of the melon PMEI gene, it was found that the length of the melon PMEI gene in between 160-252 aa, which was similar to the length of the cassava MePMEI1 protein reported so far (Zhou et al., 2019). The resemblance between the melon and ArabidopsisPMEI gene families was not high while the melon genes closely resembled those of the cucumber PMEI family was highly similar (Fig. 6). These differences may be due to differences between species. The subcellular localization of the multiple genes of the melon PMEI gene family was found to be in the cytoplasm and vacuoles. This is consistent with findings in other plants (Zhou et al., 2019). Therefore, we speculated that a low CmPMEI1 expression in the early stage of flower bud development caused demethylation of pectin, thus changing the mechanical strength and chemical properties of the cell wall, affecting fertility in the melon.
To understand the regulatory mechanisms of the MS network in melon, the AMS was used as a target gene to screen the downstream interacting partners through Chromatin Immunoprecipitation (ChIP) experiments. However, because of its low success rate in melons, no DNA fragment binding to AMS was detected in living cells. Therefore, further studies are needed to investigate the regulatory network downstream of AMS.
Conclusion
Using transcriptome sequencing and qRT-PCR, we observed the differential expression between CmZR1 and CmPMEI1 in fertile and sterile male melon plants. Yeast two-hybrid analysis showed that CmZR1 and CmPMEI1 interacted with and regulated the transcription factor AMS. We have, thus, identified a primary pathway related to male sterility in melon.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20210058_Figure_1s.pdf
Analysis of Conserved Domains of Pectinase Inhibitors and Prediction of Conserved Motifs: Phylogenetic tree analysis of melon PMEI gene family (A); Conserved Domains analysis of melon PMEI gene family (B); Prediction of conserved motif in melon PMEI gene family (C).
- HORT_20210058_Figure_2s.pdf
Determination of PME activity: 1 unit equals 1 OD600/min·g.
- HORT_20210058_Table_1s.pdf
Phylogenetic analysis of Pectin methylesterase inhibitor gene family gene list
- HORT_20210058_Table_2s.pdf
Physicochemical properties and subcellular localization of Pectin methylesterase inhibitor