Introduction
Materials and Methods
Plants Materials
Physical and Chemical Properties
Karyotype Analysis
Assessment of Genetic Diversity using RAPD and SSR Marker Analyses
Analysis of Genetic Variation using the Plastid trnL-trnF Marker
Results and Discussion
Introduction
Citrus is an economically important fruit crop through the world, and accounted for about 17% of world fruit crop production in 2018 (FAO, 2018). In Jeju, Korea, citrus is the most important commercial agricultural product in terms of area and production. Moreover, citrus plays a part in the local economy and social aspect of Jeju. In the 18th century, landrace citrus fruits, including dongjeongkyul (Citrus erythrosa Hort. ex Tan.), yoogam (C. suavissima Hort. ex Tan.), and dangyooza (DY; C. grandis L. Osb.) were utilized as offerings for the king, and at least 20 species or natural variants were reported in historic literature (Kim, 1988; Kim et al., 2001; Moon et al., 2007). However, only 12 landrace species are preserved in present-day germplasm collections (Kim et al., 2001).
Among the Korean landrace citrus types, DY is still preserved as a garden tree planted in home, gardens and backyards. It is valued by locals because of its large, edible fruits and for use in folk remedies to treat colds. In Jeju, DY has also been produced commercially and been sold for use as a nutritional supplement, tea, homeopathic medicine, and traditional food for ritualistic rites to honor ancestors. Citrus peel, especially dried citrus peel, is an essential homeopathic medicine in Oriental medicine (Kang et al., 2005). The chemical properties of Citrus spp. may account for its several medicinal uses. About 60 flavonoids are found in Citrus spp. In particular, citrus peel contains many flavanones, flavones, and polymethoxyflavones (PMFs) (Yang et al., 2019). It is well reported that PMFs in citrus are detected in the peel of citrus fruits (Nogata et al., 2006; Gattuso et al., 2007; Yang et al., 2019). The medicinal effects of flavonoids extracted from citrus include antioxidant activity (Jeong et al., 1997; Lim et al., 2006; Mokbel and Hashinaga, 2006), antimicrobial activity (Han and You, 1988), and anticancer activity (Hertog et al., 1992; Yoshimizu et al., 2004; Benavente-Garcia and Castillo, 2008). Despite the high potential of Citrus for use in medicines, particularly Korean landraces or endemic citrus, there have been few studies conducted on the taxonomy, genetics, and breeding of Korean landrace citrus as a modern breeding resource.
The phylogenic relationships of Citrus are highly complicated and ambiguous. Early taxonomic studies of the Citrus genus were mainly based on morphological and biochemical properties (Tatum et al., 1974; Barrett and Rhode, 1976; Scora, 1988). Initial molecular characterization included isozyme- (Rahman and Nito, 1994) and DNA- (Federici et al., 1998; Nicolosi et al., 2000; Abkenar et al., 2004) based analyses, which were used to more clearly elucidate the citrus taxa. Studies on genetic diversity using random amplified polymorphic DNA (RAPD) and simple sequence repeat (SSR) markers revealed a narrow genetic base within the mandarins (Citrus reticulate) (Machado et al., 1996; Coletta-Filho et al., 1998; El-Mouei et al., 2011). Furthermore, karyotype analysis of chromosomes has been widely used for determining phylogenies and speciation of Citrus spp. (Guerra, 1993; Miranda et al., 1997; Befu et al., 2000; Yamamoto and Tominaga, 2003; Carvalho et al., 2005; Yi et al., 2018a, 2018b). However, there have been few studies tracing the origin and genetic background of landrace citrus in Korea as well as Japan, neighboring the center of citrus origin.
DY has two natural variants, buk-daengyooza (BDY) and seol-daengyooza (SDY), which are distinguished according to differences in fruit characteristics. However, scientific evaluation of differences between morphological and chemical properties and the genetic differences of these natural variants has not been performed. In this study, morphological and genetic properties were evaluated among common, commercially-produced DY and its natural variants, BDY and SDY. We employed traditional classification methods and molecular phylogenetical methods, such as comparison of morphological and chemical properties of variants, chromosome karyotyping analysis, and genotyping using DNA markers, to identify genetic diversity and relationships. A better understanding of DY genetics may give some clue as to the origin of the different variants and identify potential genetic resources useful for systemic and targeted breeding programs.
Materials and Methods
Plants Materials
Three different types of DY (Citrus grandis) referred to as DY (common type), BDY (puffy type), and SDY (non-puffy type) were used in this experiments. DY was maintained at the Jeju Special Self-Governing Province Agricultural Research & Extension Service. BDY and SDY were collected from the backyards of several farmhouses in Aewol, Jeju, Korea.
Physical and Chemical Properties
To characterize physical properties, leaf length and width and the weight, length, diameter, and peel thickness of the fruit were measured. Hardness was measured three times at the equator of fruits using a penetrometer (FHM-5, Takemura, Japan). Juice was squeezed out of the fruit, and soluble solids content and acidity were measured using a soluble solids-acidity meter (GMK-707R, G-Won Hitech Co., Ltd., Seoul, Korea). Flavonoids in fruit flesh and peels were analyzed using a high-performance liquid chromatography system (e2695 Separation module) equipped with a UV-VIS detector (Waters 2489, USA). A YMC-Triart C18 column (250x4.6 mm, S-5 µm, 8 nm) was used for chromatographic separation. Ten microliters of sample was injected and delivered with the mobile phase (acetonitrile: 20mM phosphoric acid, 2:8, v/v) at the rate of 1.0 mL·min-1 and detected at 280 nm. Peak areas were quantitated with a Waters Empower System.
Karyotype Analysis
The chromosome preparation was performed according to Dutt et al. (2010) with minor modifications. Seeds were collected and germinated at 25°C in the dark, and root tips about 1 cm long were used. The root tips were excised and pretreated in 2 mM 8-hydroquinolin at 4°C for 12 h in the dark. Subsequently, the root tips were fixed in a methanol:acetic acid (3:1, v/v) solution. The fixed specimens were washed with distilled water and digested with an enzyme mixture containing 2% cellulase from Trichoerma viribe (Sigma, Japan), 1% Macerozyme R-200 (Yakult, Japan), and 0.3% Pectolyase Y-23 (Kyowa Chemical Products Co., Ltd, Japan) at 37°C for 1 h. The digested specimens were mounted on slide glasses, scattered with a drop of fixing solution using fine-pointed forceps, and air-dried. After air-drying, the preparations were counterstained with chromomycin A3 (CMA) and DAPI as described by Befu et al. (2000) with modifications. The preparations were sequentially treated with McIlvaine’s buffer (pH 7.0) containing 5 mM MgCl2 for 30 min, 0.5 mg·mL-1 CMA for 1 h, and then rinsed with McIlvaine’s buffer for 10 min. Soon after rinsing, the preparations were mounted with coverslips using Vectashield mounting medium containing 1.5 µg·mL-1 DAPI (Vector Laboratories, Burlingame, CA). Samples were observed with an epifluorescence microscope (OLYMPUS BX51, Japan) with a BV filter cassette. The images were captured using an Olympus DP71 CCD camera, analyzed using DP manager software (Olympus, Japan), and constructed using Adobe Photoshop 7.0. Five to ten cells were compared with each other for each accession and the banding patterns were determined on the basis of the number and position of CMA positive bands.
Assessment of Genetic Diversity using RAPD and SSR Marker Analyses
Total genomic DNA was extracted from fresh leaves using a Biomedic Plant gDNA Extraction Kit (Biomedic Co., Ltd., Korea; www.ibiomedic.co.kr) and quantified using a DeNovix DS-11+ spectrophotometer (DeNovix Inc., Wilmington, DE, USA). Operon primers for RAPD analysis were selected and PCR reactions were performed according to the method described in Yun (2001). Analysis of SSR markers was performed using the M13-tailed PCR method and PCR reaction and cycling conditions were performed as previously described in Schuelke (2000) and Woo et al. (2019). Fragment analysis of PCR products was performed as described previously in Kim et al. (2012). Calling of allele sizes was performed using GeneMapper software (ver. 4.0; Applied Biosystems). The shared allele frequency was calculated using PowerMarker software (v. 3.25) (Liu and Muse, 2005).
Analysis of Genetic Variation using the Plastid trnL-trnF Marker
Chloroplast DNA was extracted and the plastid trnL-trnF intergenic region was amplified using universal primers as described by Jung et al. (2005). PCR amplification was performed using an ABI 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA) in a total volume of 10 mL, containing 20 ng genomic DNA, 5 mL 2x HS Taq mix (Dongsheng Biotech, Guangzhou, China), and 0.2 mL of each 10 pmol forward and reverse primer. The conditions for PCR amplification were as follows: 5 min for initial denaturation at 95°C, 30 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C, concluding with 1 cycle of 10 min at 72°C. The PCR products were purified using Biomedic Gel & PCR Purification Kit (Biomedic Co., Ltd., Korea). The nucleotide sequences of the purified PCR product were determined using dGTP BigDye Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, CA, USA).
Results and Discussion
Measurements of phenotypic characteristics of fruit and leaves among DY (common type), BDY (puffy type), and SDY (non-puffy type) are shown in Fig. 1, Tables 1 and 2. There was no significant difference in the morphological characteristics of leaves and fruit between DY and BDY. However, the leaf and fruit shape, fruit hardness, and peel thickness of SDY were significantly different. DY and BDY had an elliptical and ovate leaf shape with an acutinate apex, while SDY had an ovate-shaped leaf with an acute apex. SDY had a petiole wing wider than those of DY and BDY. In addition, SDY produced smaller fruits than those of DY and BDY. The peel (including the rind and albedo) of SDY fruit was significantly thinner than those of DY and BDY; however, it was much harder than those of DY and BDY.
Table 1.
Morphological leaf characteristics in three different types of dangyooza (DY, dangyooza; BDY, buk-daengyooza; and SDY, seol-daengyooza)
| Varietal type |
Leaf length (mm) |
Leaf width (mm) | Leaf shape | Leaf apex shape | Size of petiole wings |
| DY | 10.4 ± 0.2 az,y | 5.1 ± 0.1 b | Elliptical | Acutinate | Small |
| BDY | 10.2 ± 0.5 a | 4.8 ± 0.3 b | Elliptical | Acutinate | Small |
| SDY | 10.5 ± 0.4 a | 6.5 ± 0.4 a | Ovate | Acute | Medium |
Table 2.
Morphological characteristics of fruit in three different types of dangyooza (DY, dangyooza; BDY, buk-daengyooza; SDY, seol-daengyooza)
| Varietal type |
Fruit weight (g) |
Fruit length (mm) |
Fruit diameter (mm) |
Fruit firmness (kg·cm-2) |
Peel thickness (mm) |
SSC (°Brix) |
Acidity (%) |
| DY | 323.0 ± 47.0 az,y | 94.5 ± 8.1 a | 93.9 ± 4.5 a | 3.16 ± 0.15 b | 12.5 ± 0.6 a | 10.8 ± 0.3 a | 3.1 ± 0.1 a |
| BDY | 336.4 ± 12.9 a | 98.7 ± 3.0 a | 96.2 ± 1.3 a | 3.52 ± 0.08 b | 11.8 ± 0.7 a | 9.0 ± 0.3 b | 3.2 ± 0.3 a |
| SDY | 240.8 ± 7.5 b | 84.0 ± 2.0 a | 79.6 ± 2.7 b | 4.33 ± 0.09 a | 8.9 ± 0.3 b | 10.5 ± 0.4 a | 3.5 ± 0.2 a |
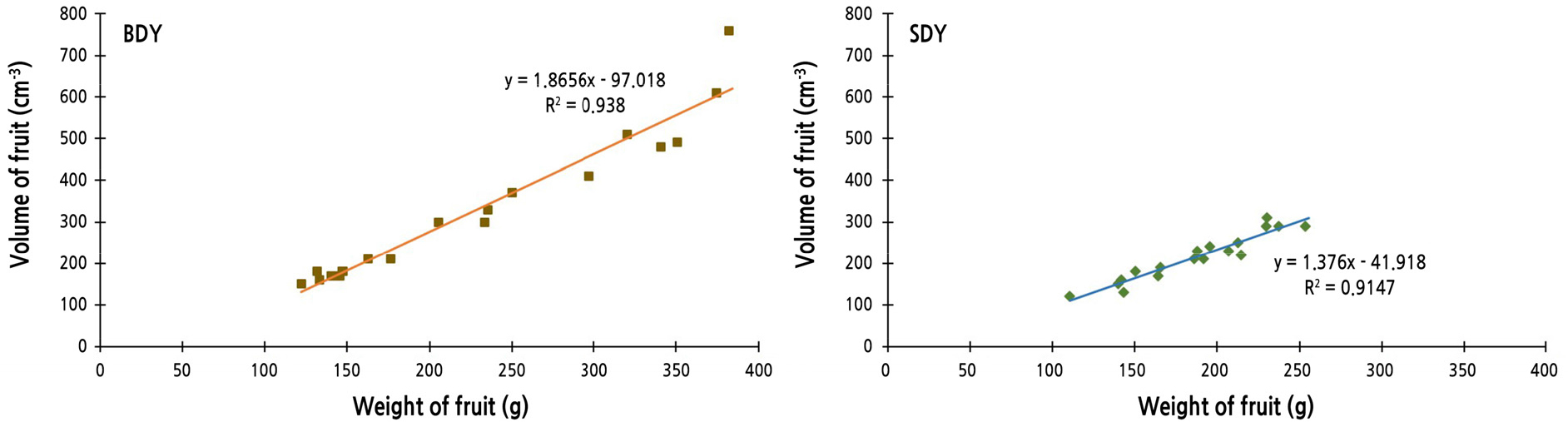
For each fruit, actual fruit volume was measured by water displacement, and weight was recorded. Linear regression equations and correlation coefficients between fruit weight and volume were calculated (Fig. 2). The value of the correlation coefficient of BDY was higher than that of SDY, indicating that fruit of BDY tends to be bigger than SDY fruit of the same weight. One of the most distinguishable characteristics of BDY and SDY is the puffiness of fruit. In fact, BDY (buk-daengyooza; “buk” means inflation or puffy in Korean) is the pseudonym or alias of DY, because the puffiness is recognizable when the fruit is pounded or pressed. In addition, the albedo layer is conspicuously thicker in DY and BDY than in SDY.
Morphological traits of citrus varieties, including fruit color, size, and weight, and leaf and fruit shape, are highly diverse. In this study, morphological traits of leaves and fruit distinguished SDY from DY and BDY, suggesting that SDY may have a different genetic background compared with DY and BDY.
Chemical properties, such as total soluble solids and acidity of citrus fruit, have been used as criteria for the classification of fruit as well, especially since these traits tend to show high heritability (Ahmed et al., 2018). Individual flavonoid concentrations in the pulp and peel (flavedo and albedo) of DY, BDY, and SDY are shown in Table 3. Most flavonoid components, except narirutin, showed higher concentrations in the peel compared with the pulp. Rutin, a commonly found flavone in citrus (Nogata et al., 2006), was detected at concentrations of about 1.8- to 2.5-times higher in the peel of DY accessions than in the pulp. The pulp of SDY contained a 55% higher concentration of rutin than those of DY and BDY. Furthermore, the highest concentration of rutin was detected in the peel of SDY, followed by DY and BDY.
Table 3.
Individual flavonoid concentration in three different types of dangyooza (DY, dangyooza; BDY, buk-daengyooza; SDY, seol-daengyooza)
| Flavonon | Pulp | Peel | |||||
| DY | BDY | SDY | DY | BDY | SDY | ||
| Rutin | 914.5 ± 36.0 bz,y | 825.2 ± 32.6 b | 1553.4 ± 56.5 a | 2320.3 ± 120.2 b | 1605.5 ± 65.7 c | 2814.4 ± 129.3 a | |
| Narirutin | 165.3 ± 6.9 b | 213.8 ± 7.7 a | 231.8 ± 9.2 a | 191.1 ± 11.0 b | 242.9 ± 9.3 a | 188.3 ± 8.9 b | |
| Naringin | 2126.3 ± 95.6 b | 1644.3 ± 61.6 c | 2896.0 ± 111.2 a | 3234.5 ± 166.7 a | 3028.6 ± 112.8 a | 3532.7 ± 159.2 a | |
| Hesperidin | 32.7 ± 2.7 a | 36.8 ± 1.4 a | 77.5 ± 3.6 b | 108.7 ± 6.2 a | 104.7 ± 4.6 a | 155.1 ± 10.3 b | |
| Neohesperidin | 1225.7 ± 59.5 b | 1134.2 ± 44.1 b | 2595.7 ± 105.6 a | 3445.1 ± 181.8 b | 2654.9 ± 104.9 c | 4547.2 ± 210.2 a | |
| Nobiletin | - | - | - | 113.2 ± 4.2 a | 113.5 ± 4.6 a | 117.5 ± 3.9 a | |
| Tangeretin | - | - | - | 68.8 ± 1.9 a | 62.5 ± 1.4 b | 57.4 ± 1.0 b | |
The major flavanones in citrus, including naringin, hesperidin, and neohesperidin, were also higher in both the pulp and peel of SDY than in DY and BDY. Narirutin was higher in only the peel of SDY. The concentrations of hesperidin and neohesperidin in the pulp and peel of SDY were considerably higher than those of DY and BDY. Moreover, the pulp of SDY contained almost twice as much hesperidin and neohespiridin. The two major PMFs in citrus, nobiletin and tangeretin, were only detected in the peel of all three types of DY, and there were no significant differences in their concentrations.
Chemical composition studies, in addition to morphological characteristics, were applied to the taxonomic classification of citrus, and it has resulted in remarkable progress in citrus taxonomic studies (Barrett and Rhodes, 1976). For instance, nucellar seedlings could be distinguished from zygotic seedlings by methoxy flavonoid compounds (Tatum et al., 1974).
The somatic metaphase chromosomes of BDY and SDY were all diploid (2n = 18). Banding patterns from CMA staining of chromosomes were typed based on the number and position of CMA positive bands according to Befu et al. (2000) and Miranda et al. (1997) (Fig. 3). The chromosome configurations of both BDY and SDY were 1A + 3B + 1C + 7D + 6E, which is identical to that of DY (Yi et al., 2018b). Both BDY and SDY possessed a total of five A-, B- and C-type chromosomes. Such large numbers of A, B, and C chromosomes are typical for CMA banding patterns commonly found in C. maxima (Guerra, 1993; Miranda et al., 1997; Befu et al., 2001). Due to its self-incompatibility and broad cultivation worldwide, pomelo (C. maxima) can be easily hybridized to produce diverse cultivars, varieties, and strains. However, the result of this study revealed that there was no variation in chromosome composition among DY, BDY, and SDY. This suggests that they are not distinct hybrids or strains resulting from interspecific hybridization.
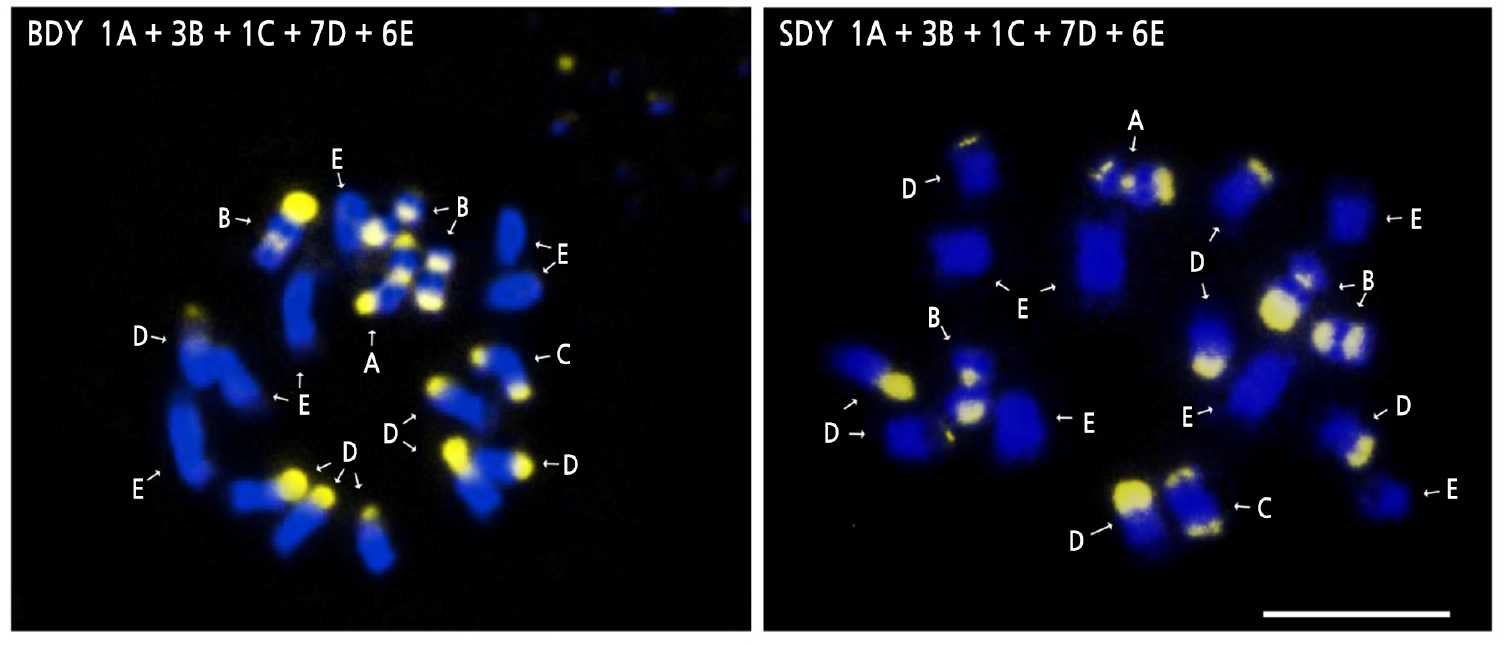
Fig. 3.
CMA/DAPI-stained somatic chromosomes in buk-daengyooza (BDY) and seol-daengyooza (SDY). Letters indicate the number and distribution of CMA positive bands as follows; A, two telomeric and one proximal band; B, one telomeric and one proximal band; C, two telomeric bands; D, one telomeric band; and E, no band. The gray regions signify CMA positive bands. Scale bar indicates 5 µm.
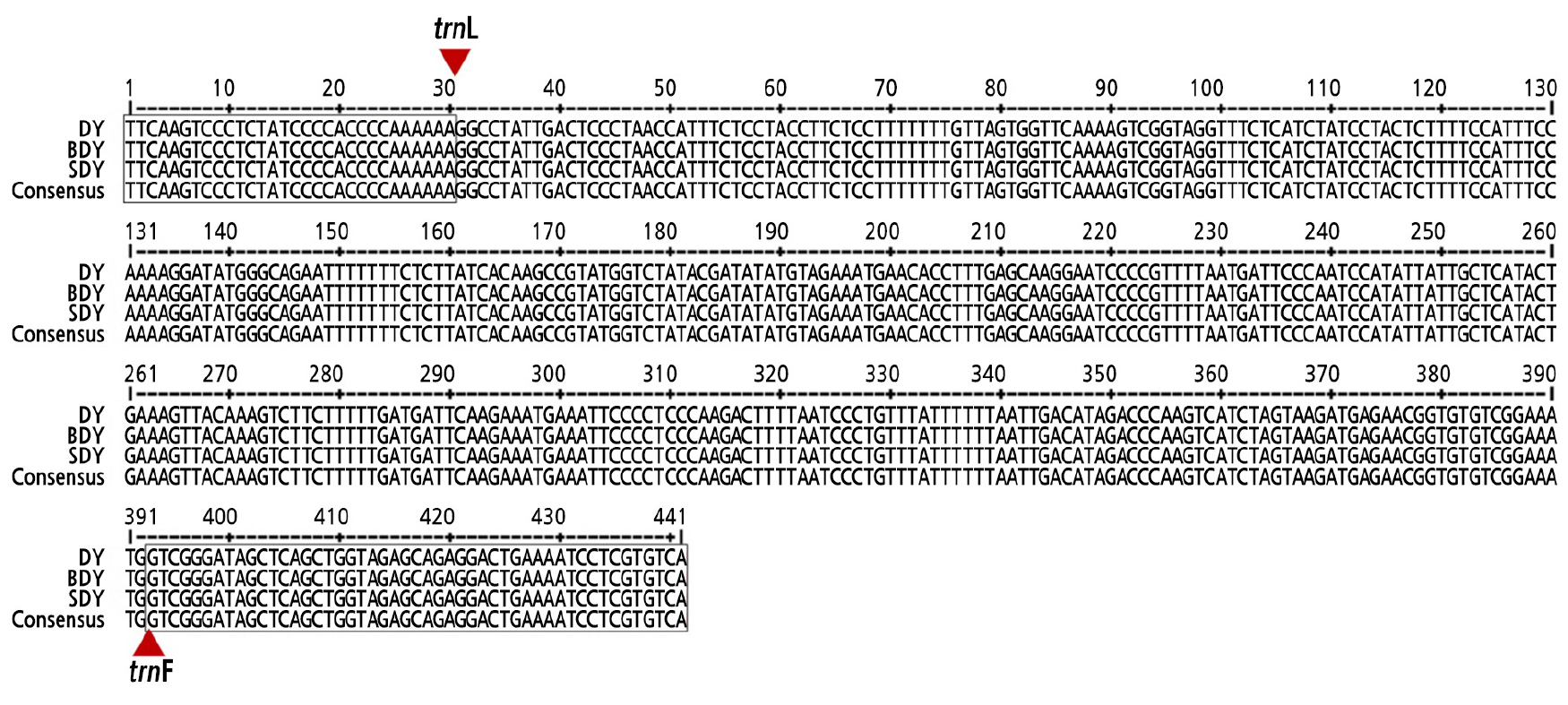
The genetic variation and diversity among DY, BDY, and SDY was analyzed to distinguish them at a molecular level using RAPD, SSR, and trnL-trnF barcoding markers. However, there was no polymorphism found among the three accessions. No polymorphic bands were revealed by the RAPD and SSR data (Suppl. Fig. 1s and Suppl. Table 1s). Nucleotide sequences of the amplified trnL-trnF intergenic region were identical among the three accessions without any different SNPs (Fig. 4). Given these results, DY, BDY, and SDY are all very closely related.
DNA barcoding markers, such as trnL-trnF, are considered powerful tools for identifying field gametophytes (Chen et al., 2013). These markers have been widely used for studying phylogenetic relationships in Citrus (Jung et al., 2005; Yingzhi et al., 2007; Jena et al., 2009; Li et al., 2010; Lu et al., 2011). Genetic diversity and phylogenetic relationships among many citrus cultivars have been analyzed using RAPD markers (Machado et al., 1996; Coletta-Filho et al., 1998; Federici et al., 1998; Abkenar and Isshiki, 2003; Baig et al., 2009) and SSR markers (Biswas et al., 2011; Shrestha et al., 2012; Nematollahi et al., 2013; Ahmed et al., 2018). SSR markers have also been applied for citrus germplasm collection (Barkley et al., 2006) and identification of zygotic and nucellar seedling in citrus (Yildiz et al., 2013; Woo et al., 2019).
In this study, we analyzed phenotypic, phytochemical, cytogenetic, and genotypic characteristics of DY, BDY, and SDY to elucidate their phylogenetic relationship. Morphological and phytochemical traits of leaves and fruit distinguished SDY from DY and BDY. The karyotype analysis revealed that there was no polymorphism among the CMA/DAPI banding patterns for the accessions tested. Despite morphological and phytochemical differences among DY, BDY, and SDY, chromosomal configuration and DNA marker analyses were unable to clearly discriminate their phylogenetic relationships. This suggests that SDY and BDY may have differentiated from DY with highly similar genetic backgrounds. The distinct morphological and phytochemical traits of SDY may have also arisen from a somatic mutation, such as a nucellar seedling.
The data and results from this study lay groundwork for future study of DY. By having a better understanding of morphological, chemical, and genetic differences among DY, BDY, and SDY, continued experiments can be designed and used to identify potential genetic resources useful for systemic and targeted breeding programs. Furthermore, these findings give some preliminary clues to the origin of the different variants.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20210009_Fig_1s.pdf
PCR amplified products of 28 RAPD primers with dangyooza (DY), buk-dangyooza (BDY), and seol-dangyooza (SDY) accessions (L1; DY, L2; BDY, and L3, SDY).
- HORT_20210009_Table_1s.pdf
Allele sizes of dangyooza (DY), buk-daengyooza (BDY), and seol-daengyooza (SDY) accessions determined by 24 polymorphic SSR markers