Introduction
Materials and Methods
Plant Material
Multiplication Stage
Rooting Stage
Molecular Marker Analysis for Genetic Stability Assessment
Statistical Analysis
Results
Multiplication Stage
Rooting Stage
Molecular Marker Analysis for Genetic Stability Assessment
Discussion
Conclusion
Introduction
Olive (Olea europaea L., Family: Oleaceae) is one of the traditionally cultivated fruit crops in the Mediterranean basin and has a vital economic role in Mediterranean countries (Baldoni and Belaj, 2009). The olive’s geographic origin goes back to the Mediterranean basin’s eastern coast (Connor, 2005). Olive trees are well adapted to the semi-arid environment and cultivated under rain-fed conditions without needing supplemental irrigation (Connor and Fereres, 2005; Dag et al., 2008). Olive fruits, oil, and leaves are rich in valuable bioactive pharmaceutical materials, and the benefits of olive products to human health are numerous and broadly recognized (Visioli et al., 2002; Ghanbari et al., 2012). A growing awareness of olive oil’s nutritional value has boosted the international demand for olives and olive oil (Torres et al., 2017). Olive trees are propagated by leafy stem cuttings under mist conditions; the rooting ability depends on the genotype and environmental conditions (Hartman et al., 2007). Cultivars that are difficult to root are reproduced by grafting on seedlings or clonal rootstocks (Sotomayor-León and Caballero, 1994; Fabbri et al., 2004). Several studies have addressed the limitations of the traditional propagation methods of olive trees (Rkhis et al., 2011). Micropropagation is a powerful technique for propagating olive trees in high quantities, specifically the hard-to-root genotypes. Micropropagation is also a valuable tool for genetic improvement and germplasm conservation with pathogen-free and genetically uniform plant materials (Zuccherelli and Zuccherelli, 2002; Zacchini, and De Agazio, 2004). The following factors can affect micropropagation: plant genotype, physiological status of mother plant, explant type and age, mineral content of culture media, carbon source, and growth regulators (Grigoriadou et al., 2002; Zuccherelli and Zuccherelli, 2002; Peixe et al., 2007; Leva et al., 2013).
Media browning caused by the oxidation of phenolic compounds, strong apical dominance, and slow lateral olive shoot growth, are the primary problems of olive micropropagation (Rugini, 1984; Lambardi et al., 2012; Benelli and De Carlo, 2018). Growth regulators are critical elements in the in vitro culture media; cytokinin type and concentration substantially affect shoot growth and multiplication rate. Moreover, using unsuitable types or concentrations of cytokinins may adversely affect the shoot growth and proliferation rate of different plant species (Rugini, 1984; Peixe et al., 2007; George et al., 2008; Lambardi et al., 2012). Zeatin, the most utilized cytokinin, induces a satisfactory growth and proliferation rate of in vitro cultured olive explants (Rugini, 1984; Rugini, 1990; Grigoriadou et al., 2002; Rugini and Baldoni, 2004). The replacement of zeatin with other synthetic cytokinins, such as 6-benzylaminopurine, thidiazuron, and kinetin, yielded unsatisfactory results with most olive cultivars (Briccoli-Bati and Lombardo 1995; Mencuccini et al., 1997; Dimassi, 1999; Roussos and Pontikis, 2002). In vitro root formation is critical during woody plant micropropagation. Many endogenous and exogenous factors affect the in vitro rooting ability, including the genotype’s rooting potential, the mother plant’s age, environmental conditions, nutrient media, and the type and concentration of auxin (Rugini and Pannelli, 1993). Auxins, specifically IBA and NAA, have been used to root olives (Jimenez, 2005; Tanimoto, 2005). However, the type and concentration depend on the olive cultivar (Soumendra et al., 2000; Grigoriadou et al., 2002).
The start codon targeted (SCoT) marker is a reliable marker system that can be used to preferentially detect polymorphisms in the coding sequences of some horticultural species, such as mango (Luo et al., 2012; Zhou et al., 2020), olive (Alsamman et al., 2017) and pear (Jalilian et al., 2018). However, few studies have used the SCoT marker to determine the genetic diversity and relationships among olive cultivars in Egypt (Mohamed et al., 2017). Therefore, this study aimed to optimize olive shoot propagation by evaluating olive growth in response to different zeatin concentrations and assessing the genetic stability of olive plants produced under in vitro conditions.
Materials and Methods
Plant Material
The current research was conducted in a laboratory at the Pomology Department, Faculty of Agriculture, Cairo University, Giza, Egypt. Active spring shoots were collected from mature ‘Coratina,’ ‘Picual,’ ‘Frantoio,’ ‘Manzanillo,’ ‘Toffahi,’ and ‘Aggizi Shami’ olive trees grown at the experimental olive orchard (situated at 031°12'65"E longitude and 30°00'48"N latitude). Shoots were stripped of leaves, washed with tap water, and divided into nodal cuttings. The cuttings were surface sterilized with commercial bleach (5.25% sodium hypochlorite) for 10 min, followed by mercury chloride at 1000 mg·L-1 for 5 min, and then washed several times with sterile distilled water.
Multiplication Stage
Olive nodal cuttings were cultured on Rugini Olive media (OM) (Rugini, 1984) supplemented with zeatin (6-γ,γ- dimethylallyl-amino) at 2, 4, and 6 mg·L-1, 30 g·L-1 mannitol, and 6 g agar L-1. The pH of the media was adjusted to 5.8 before adding agar, and then the media was autoclaved at 121°C for 15 min. All cultures were kept in a growth chamber at 25 ± 2°C with a 16 h photoperiod. Five weeks later, the sprouted buds were transferred to fresh media with the same composition, and a subculture was performed every five weeks. The proliferation rate, shoot length, node number per shoot, internode length, and leaf number were recorded at the end of each culture period.
Rooting Stage
Healthy shoots from the 3rd subculture were used to determine root induction. In vitro regenerated micro-shoots with uniform sizes from different olive cultivars were selected. Two rooting methods were explored. The single-phase rooting method involved culturing olive shoots on ½OM supplemented with 2 mg·L-1 of indole-3-butyric acid (IBA). The two-phase rooting method involved a quick-dipping treatment (30 seconds) of the shoot base in a sterile IBA solution at 250 mg·L-1. The shoots were then transferred to ½OM without hormones (Liu and Pijut, 2008). All media were supplemented with 30 g·L-1 mannitol and 6 g agar L-1. The rooting media were darkened with 1 g·L-1 active charcoal. Two months later, the rooting percentage, number of roots per shoot and root length (cm) were recorded.
Molecular Marker Analysis for Genetic Stability Assessment
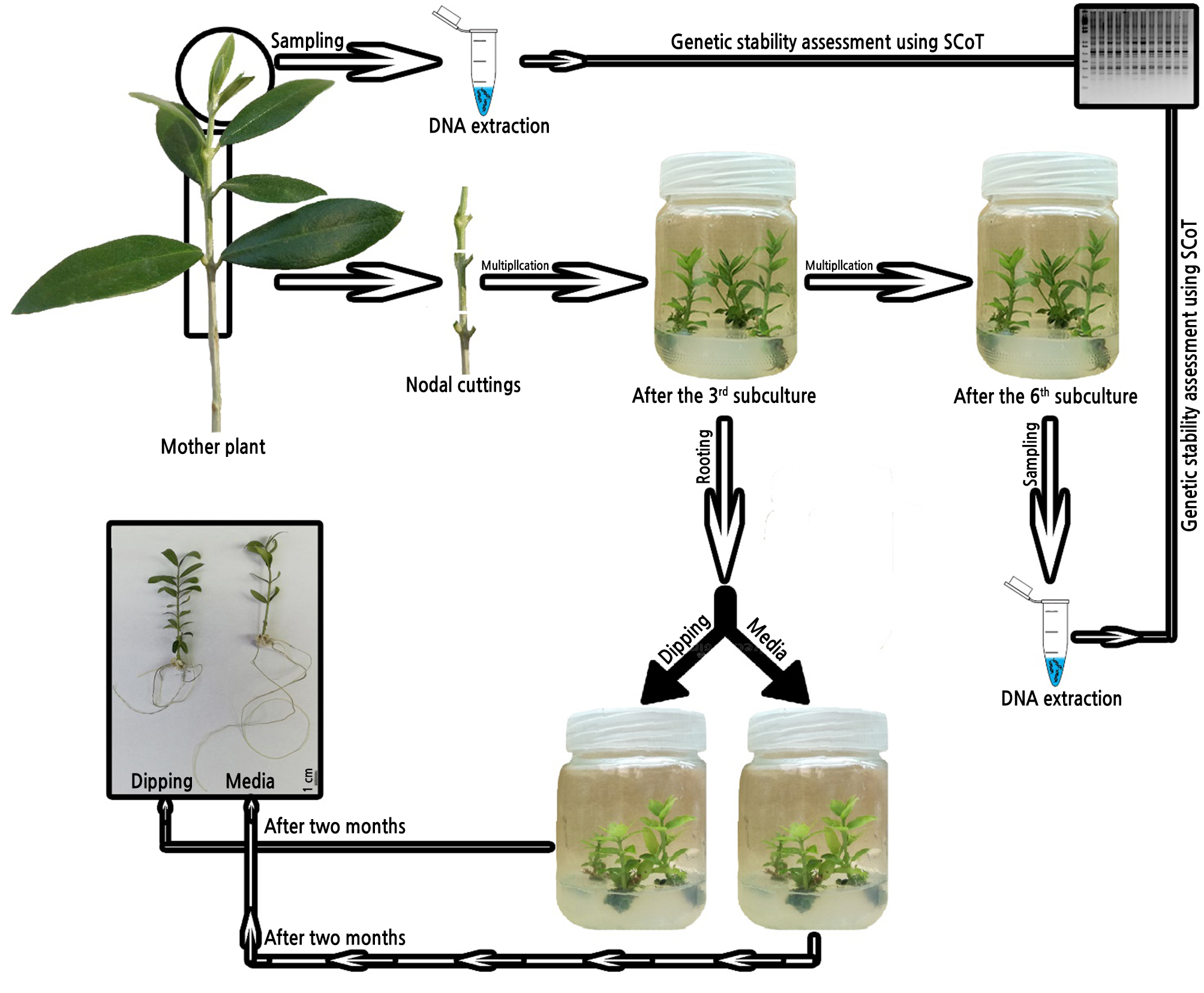
Genomic DNA was extracted from fresh leaves of each cultivar—from the 6th subculture and from mother plants—using a DNeasy plant mini kit (Qiagen, USA). The DNA quality was checked by determining the absorbance ratio at A260/A280 with a UV-spectrophotometer (Apel, PD-303UV, Japan), where DNA is pure at an A260/A280 ratio from 1.8 to 2.0. DNA quality was checked using electrophoresis (Fotodyne 300, USA) in a 1% agarose gel with ethidium bromide. SCoT amplification was performed as described by Collard and Mackill (2009) using seven primers (Table 1) selected from our laboratory’s SCoT primers bank. These primers were developed from a consensus sequence derived from previous studies by Joshi et al. (1997); Sawant et al. (1999); Collard and Mackill (2009); and Xiong et al. (2011) and procured from Biobasic Com. All SCoT primers were 18-mer and were from Dataset I, which is based on highly expressed genes described by Sawant et al. (1999). Amplification reactions for SCoT techniques were performed as specified by Xionget al. (2011) and were conducted in a Thermal Cycler (Techne TC-512, USA) as follows: 94ºC for 4 min; 40 cycles of 1 min at 94ºC, 1 min annealing at 57ºC, and 2 min at 72ºC; and 72ºC for 10 min. The reaction products were stored at 4ºC. Amplified products were loaded and separated on a 1.5% agarose gel with ethidium bromide and 100 bp to 1.5 kb ladder markers for 30 min at 100 V in a horizontal gel apparatus (Mini-submarine gel Bio-Rad, USA). DNA banding pattern photos were acquired using a gel documentation system (Bio-Profil, Bio-1D, USA) and analyzed by GelAnalyzer 3 19.1 software, scoring clear amplicons as present (1) or absent (0) for each primer and entering the results in a binary data matrix. Molecular distances were determined by the Nei and Li coefficient (Nei and Li, 1979) utilizing the binary data matrix. Similarity matrices were produced using Gel works ID advanced software (UVP-England Program). The DICE computer package was used to produce a pairwise difference matrix (Yang and Quiros, 1993). A graphical representation scheme of all steps done in this work, including multiplication, rooting, and genetic stability assessment, is shown in Fig. 1.
Table 1.
List of the primers and their nucleotide sequences used in the study for the SCoT procedure
Statistical Analysis
Each treatment had three biological replicates (seven jars per replicate) with four explants in each culture jar. The experiment was carried out in a randomized complete block design (Snedecor and Cochran, 1967). The assumptions of normality were tested using Shapiro-Wilk's test (Shapiro and Wilk, 1965). The homogeneity of variance between different subcultures was assessed using Levene’s test (Levene, 1960). Two-way analyses of variance (ANOVAs) were used to determine the effects of genotype, zeatin concentration, and their interaction. ANOVAs were performed using the SAS software (version 9.0; SAS Institute, Cary, NC). The means were calculated from three replicates per treatment and the significant differences within and between treatments were evaluated with multiple Duncan range tests at a significance level of 0.01 (Duncan, 1955). A linear regression model was employed to describe the changes in growth parameters of olive shoots grown in vitro after six subcultures.
Results
Multiplication Stage
Both plant genotype and zeatin concentration had a significant effect (p < 0.01) on the number of shoots (Table 2). Differences in shoot number existed between the different investigated olive genotypes; the ‘Toffahi’ and ‘Aggizi Shami’ cultivars had a significantly higher number of shoots (1.841 and 1.945, respectively) compared with the other cultivars. The number of shoots increased with increasing zeatin concentration in the OM. The maximum number of shoots (1.804) was recorded at a zeatin concentration of 6 mg·L-1. Regarding the interaction between olive genotype and zeatin concentration, the highest number of shoots was recorded for ‘Toffahi’ and ‘Aggizi Shami’ (2.085 and 2.208, respectively) growing on OM supplemented with zeatin at 6 mg·L-1, while the lowest response was reported for ‘Picual’ and ‘Coratina’grown on OM with zeatin at 2 mg·L-1.
Table 2.
Effects of zeatin concentration on shoot number of different olive cultivars
Table 3 shows that shoot and internode length markedly varied among the studied olive cultivars; with the highest shoot length (6.166 cm) in ‘Coratina,’ followed by ‘Frantoio’ (5.411 cm) and ‘Picual’ (5.294 cm), while ‘Manzanillo’ had the lowest value (4.536 cm). Zeatin concentration had a slight but non-significant effect on shoot elongation. The highest internode length (1.297 cm) was recorded for ‘Aggizi Shami,’ and the other cultivars had similar internode lengths. Increasing zeatin concentration in the OM slightly reduced the internode length; however, there were no significant differences among the three investigated zeatin concentrations.
Table 3.
Effects of zeatin concentration on the shoot and internode length of different olive cultivars
Both genotype and zeatin concentration had a significant effect (p < 0.01) on the number of leaves and the number of nodes per shoot (Table 4). ‘Coratina’ and ‘Frantoio’had the highest numbers of leaves and nodes per shoot, followed by ‘Picual’, while ‘Aggizi Shami’ had the lowest values. Zeatin at 6 mg·L-1 resulted in a significantly (p < 0.01) higher number of leaves (10.35) and nodes (5.177) per shoot compared with other concentrations. The highest number of leaves and nodes per shoot was recorded for ‘Coratina’ shoots growing on OM supplemented with zeatin at 6 mg·L-1, while the lowest number of leaves and nodes per shoot was recorded for ‘Aggizi Shami’ growing on OM supplemented with zeatin at 2 mg·L-1.
Table 4.
Effects of zeatin concentrations on the number of nodes and leaves per shoot of different olive cultivars
Table 5.
Linear regression analysis for growth parameters of in vitro-grown olive shoots over 6th subcultures
A linear regression analysis was conducted to estimate the changes in olive shoot growth parameters after the sixth subculture (Table 5). It revealed that the shoot number increased linearly with time. The slope (β1) was similar for ‘Coratina,’ ‘Picual,’ ‘Frantoio,’ and ‘Manzanillo’, but lower for ‘Toffahi,’ (β1 = 0.005778). The high correlation coefficient indicated a strong positive relationship between shoot number and the number of subcultures. However, regression analysis revealed that shoot length decreased linearly with time in ‘Coratina,’ ‘Picual,’ ‘Frantoio,’ and ‘Manzanillo’, with negative slope (β1) values. The positive correlation coefficient indicated a positive relationship between shoot length and the number of subcultures in ‘Toffahi’ and ‘Aggizi Shami’. The results confirm that subculture frequency affected in vitro olive shoot growth behavior.
Rooting Stage
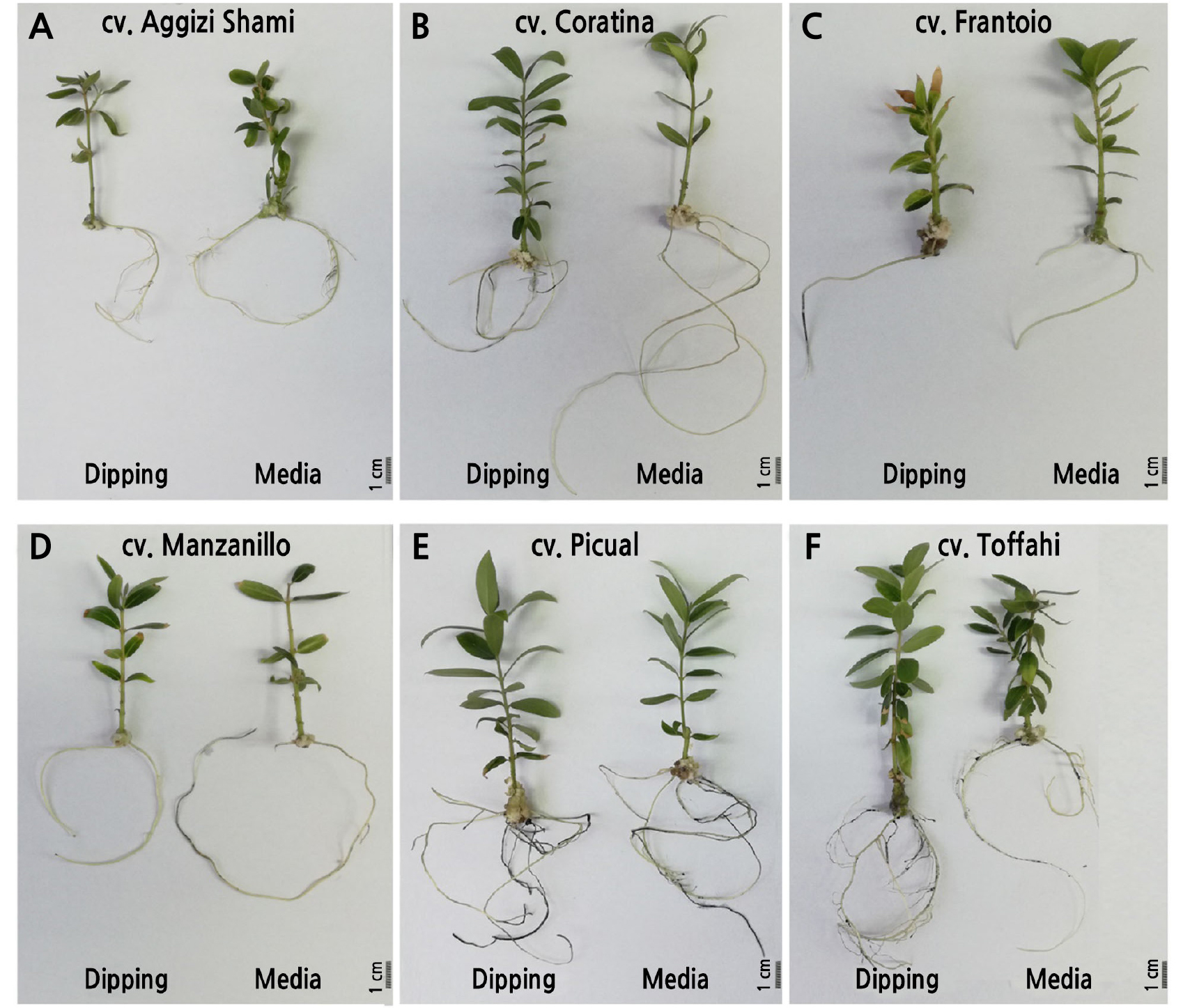
The effects of genotype and rooting method on olive micro-shoot rooting is illustrated in Table 6 and Fig. 2. The highest rooting percentage was recorded for ‘Coratina’ (Fig. 2B; 67.87%), followed by ‘Picual’ (Fig. 2E; 53.34%), while the lowest rooting percentage (12.83%) was recorded for ‘Aggizi Shami’ (Fig. 2A). The number of roots and root length were genotype-dependent traits. The ‘Picual’ cultivar had the highest number of roots per shoot (3.633) and root length (13.72 cm), while the ‘Frantoio’ cultivar had the lowest (1.463 and 6.775 cm, respectively). There were significant differences (p < 0.01) between the rooting treatments for rooting percentage, root length, and the number of roots per shoot. Olive shoots grown on OM with 2 mg L-1 IBA had a higher rooting percentage (45.637%) compared with shoots subjected to the two-phase dipping method (34.53%); the highest rooting percentage was recorded for the ‘Coratina’ cultivar with the dipping method (71.08%). The medium with 2 mg·L-1 IBA resulted in a significantly higher (p < 0.01) root length, while the two-phase dipping method resulted in a significantly higher (p < 0.01) number of roots per shoot.
Table 6.
Effect of rooting method on the rooting percentage, number of roots per shoot, and root length (cm) of different olive cultivars
Values of interaction (treatment × cultivar) followed by different lowercaseletters are significantly different (p < 0.01). Mean values of treatments followed by asterisks are significantly different (p < 0.01). Mean values of cultivars followed by different uppercaseletters are significantly different (p < 0.01).
Molecular Marker Analysis for Genetic Stability Assessment
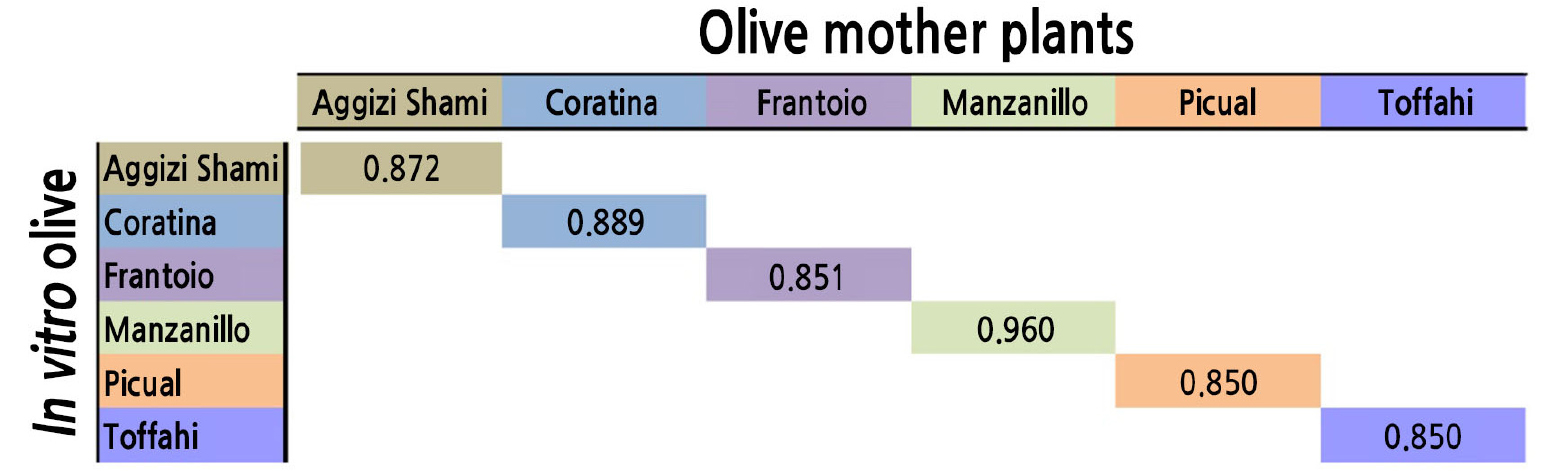
The SCoT marker technique was used to determine genetic stability and the presence of somaclonal variation in tissue-derived olive plants over successive subcultures. The highest similarity value (between an in vitro tissue-derived plant and its mother plant) was recorded for ‘Manzanillo’ (0.960), followed by ‘Coratina’. This result indicates that these two cultivars exhibited high genetic stability during the in vitro propagation over the six subcultures. The lowest values were reported for ‘Picual’ and ‘Toffahi’ (0.850 for both), indicating that these two cultivars were less genetically stable during the in vitro propagation (Fig. 3).
Discussion
Olive is one of the most economically important fruit crops in the Mediterranean basin. A growing awareness of the nutritional value of olive oil has increased the international demand for olives and olive oil (Torres et al., 2017). Hence, improving plant propagation procedures to overcome the limitations of traditional olive tree propagation methods is exceedingly crucial for the olive industry (Rkhis et al., 2011). The results obtained here indicated that the cultivar and cytokinin concentration are important factors in olive micropropagation. Increasing the zeatin concentration in the culture media increased the number of shoots and the number of internodes per shoot. As olive shoots exhibit strong apical dominance, axillary shoot formation is limited. Thus, multiplication is achieved by shoot segmentation at each subculture (Lambardi and Rugini, 2003; Fabbri et al., 2004). The shoot multiplication rate relies primarily on the number of shoots and nodes. Therefore, a higher number of shoots and internodes allows increased proliferation and therefore higher micropropagation rates.
Increasing zeatin levels resulted in a reduction in shoot length, demonstrating a negative effect of zeatin on shoot length. The olive shoot internode elongation rate was slightly affected by zeatin concentration, and higher zeatin concentrations reduced shoot elongation (Haddad et al., 2018). In vitro propagation of olive is dependent on cytokinin type and concentration, and the results obtained here align with the previous study by Grigoriadou et al. (2002). Cytokinins are crucial components of the in vitro culture media used during the multiplication stage, reducing shoot apical dominance, and stimulating basal shoot formation (Fabbri et al., 2004; Panjaitan et al., 2007). The effect of cytokinin on shoot number may be related to its inhibition of apical dominance, stimulating shoot formation, and reducing shoot length (Huetteman and Preece, 1993; Malik et al., 2005). Our results showed that zeatin concentration plays an essential role in the in vitro propagation of olive. Zeatin is the most effective cytokinin to enhance shoot regeneration in a broad range of olive cultivars (Grigoriadou et al., 2002; Rugini and Baldoni, 2004; Ali et al., 2009; Rugini et al., 2016).
Moreover, the stability of repetitive subcultures is critical in plant tissue culturing to avoid somaclonal variation (Leva et al., 2012; Weckx et al., 2019). The effect of repeated subculturing on the growth of in vitro shoots has previously been reported (Norton and Norton 1986; Debnath, 2004; Lo et al., 2012; Vujović, et al., 2012). Its impact on the multiplication rate of in vitro-grown shoots depends on the plant genotype (Vujović, et al., 2012). The shoot multiplication rate gradually declined during long-term subculture in six Rosaceae species (Norton and Norton, 1986). In contrast, shoot multiplication increases with subculturing in cherries, apples, pineapple, and raspberry (Grant and Hammat, 1999; Debnath, 2004; Hamad and Taha, 2008). The increase in shoot number over an extended subculturing time was attributed to plant tissue rejuvenation during in vitro culturing (Grant and Hammat, 1999).
Rooting of in vitro-generated shoots is critical for any successful micropropagation protocol (Blazkova et al., 1997; Caboni et al., 1997). In olives, the rooting ability depends on the plant genetic background, media composition, auxin type and concentration, and incubation conditions (light and temperature) (Rugini, 1990; Benelli et al., 2001; Grigoriadou et al., 2002). Rooting ability varies among olive cultivars, and in vitro rooting has been unsuccessful for some olive cultivars (Dimassi-Theriou, 1994; Briccoli-Bati et al., 1999; Rugini et al., 1999). The results obtained here show that rooting potential differs significantly between olive cultivars. These results aligned with those of Rugini (1984); Fiorino and Leva (1986); and Rama and Pontikis (1990), who reported that in vitro rooting ranged from 25 to 85%, depending on the cultivar.
Exogenous auxin applications are vital in micro-shoot rhizogenesis (Soumendra et al., 2000; Grigoriadou et al., 2002). Auxin type and concentration strongly influence the quality of regenerated roots (Jimenez, 2005; Tanimoto, 2005; Ali et al., 2009). IBA is the most effective auxin for olive rhizogenesis (Krieken et al., 1993; Liu et al., 1998; Benelli et al., 2001; Tanimoto, 2005). Here, two in vitro rooting methods were explored; the traditional single-phase rooting method (Soumendra et al., 2000; Grigoriadou et al., 2002; Tanimoto, 2005) and the two-phase or “root dipping” method, in which the base of micro-shoots is quickly dipped in a concentrated IBA solution (150–250 mg·L-1) before culturing in a hormone-free medium (Bartolini et al., 1990; Rugini and Fedeli, 1990). The single-phase rooting method (on ½OM with 2 mg·L-1 IBA) resulted in a higher rooting percentage and root length compared with the double-phase rooting method. This result is in contrast with that of Rugini and Fedeli (1990), who reported high rooting percentages when olive micro-shoots were dipped in an IBA solution. Based on shoot growth and rooting potential, our results indicate that ‘Coratina’ and ‘Picual’ are the most adaptable cultivars to the in vitro conditions, followed by ‘Frantoio’ and ‘Manzanillo’. However, we consider ‘Aggizi Shami’ and ‘Toffahi’ to be recalcitrant cultivars to in vitro conditions, as they exhibited a low rooting potential.
Furthermore, the genetic stability of the six cultivars was evaluated by analyzing the SCoT markers in tissue-derived plants and parental material. The results indicated that repetitive subculturing negatively affects in vitro olive micropropagation. These results contrasted with previously published data by Hassan et al. (2016) and Leva and Petruccelli (2012), who used the RAPD marker to evaluate the genetic stability of several micropropagated olive cultivars and reported that in vitro olive micropropagation did not affect morphological characterization or genetic stability. This discrepancy may be due to the difference in the sampling time for molecular analysis and the number of subcultures.
Conclusion
The performance of in vitro olive propagation usually depends on the genotype and growth regulators. By increasing the zeatin concentration, the proliferation rate increased. Additionally, the shoot number of all studied olive cultivars increased linearly with time. The addition of indole-3-butyric acid (IBA) to culture media exhibited better results than the dipping method. ‘Coratina’ and ‘Picual’ are the most adaptable cultivars to the in vitro conditions, followed by ‘Frantoio’ and ‘Manzanillo.’ ‘Aggizi Shami’ and ‘Toffahi’ are recalcitrant cultivars to the in vitro conditions, as they demonstrated a low rooting potential. The repeated subculture negatively affected the genetic stability of all cultivars. The genetic similarity results from the SCoT marker data indicated that the SCoT technique is viable for discriminating and identifying olive cultivars and can be efficiently used for future simplified molecular assessment of olive germplasm. In summary, a new method for propagating olives is required to meet future market demands. This study lays the foundation for optimizing in vitro olive propagation in Egypt. Future studies should explore this protocol in other olive cultivars grown in different Egyptian districts.