Introduction
Materials and Methods
Plant materials and germination of yuzu seeds
DNA extraction and HRM marker analyses
Constructing a linkage map of HRM markers and identifying sweet orange orthologs
Results
Identification of yuzu seedlings originating from zygotic embryos using molecular markers
Genotyping of the CitRWP gene controlling nucellar embryony and determination of the optimal set of HRM markers based on linkage relationships
Discussion
Identification of yuzu seedlings originating from zygotic embryos using molecular markers, as well as the morphological and genetic features of zygotic seedlings
Development of an optimal set of HRM markers for selecting zygotic seedlings and their application in yuzu breeding
Introduction
Yuzu (Citrus junos Sieb. Ex Tanaka), originating from China, was introduced to Japan and Korea over 1,000 years ago (Taninaka et al., 1981). It has since become one of the most important Citrus crops in Korea. The most conspicuous feature of yuzu is its delightful fruit fragrance. However, due to its intensely tart flavor, yuzu is generally consumed in processed forms, such as in beverages, perfumes, and oils (Matsumoto et al., 2016; González-Mas et al., 2019; Choi et al., 2022). Yuzu is also a key ingredient in many food products. In addition, research has shown that yuzu offers several health benefits, including antioxidant and anti-inflammatory effects (Hirota et al., 2010; Yu et al., 2011; Kim et al., 2013; Lee et al., 2020).
Although extensive therapeutic and nutritional studies have concentrated on yuzu, the availability of its genetic and genomic information is relatively limited compared to other major Citrus crops such as sweet orange (Kim et al., 2010; Ghorabaie et al., 2010). Since the initial draft of the whole-genome sequence (367 Mb) of sweet orange was published (Xu et al., 2013), extensive genome sequence analyses of primitive, wild, and cultivated Citrus species have shed light on the origins and domestication history of these species (Wu et al., 2014, 2018, 2021; Wang et al., 2017, 2018; Rao et al., 2021).
The origin of yuzu has been a topic of controversy. Tanaka (1954, 1969) initially claimed that yuzu was not a hybrid and assigned it a scientific name. Several studies (Hirai et al., 1986; Uzun et al., 2009; Shimizu et al., 2016) supported Tanaka’s hypothesis through analyses of molecular markers. However, Swingle and Reece (1967) proposed that yuzu could be a natural hybrid between Ichang papeda (C. cavaleriei) and mandarin (C. reticulata). Recently, more extensive genome-wide single nucleotide polymorphism (SNP) analyses of yuzu and related Citrus species have suggested that yuzu is most likely a natural hybrid between wild mandarin and Ichang papeda (Demarcq et al., 2021; Wu et al., 2021; Jeong et al., 2022).
During the domestication of Citrus species, apomictic reproduction has played a crucial role in the development of modern Citrus varieties (Wang et al., 2017; Wu et al., 2021). Among the various types of apomixis, nucellar embryony, by which embryos develop from the somatic nucellar tissue surrounding the embryo sac, is widespread in many Citrus crops, including yuzu, sweet orange, mandarin, and lemon (Xu et al., 2021). Due to the development of multiple embryos from nucellar tissues, nucellar embryony is also known as polyembryony (Koltunow et al., 1996; Zhang et al., 2018; Xu et al., 2021). The CitRWP gene, which regulates egg-cell-related genes, has been identified as the causal gene for nucellar embryony in Citrus (Wang et al., 2017; Shimada et al., 2018). The insertion of miniature inverted-repeat transposable elements (MITE) into the promoter region resulted in the overexpression of CitRWP,leading to nucellar embryony. Four mutant alleles containing two inverted or three tandem MITE insertions have been observed in various Citrus species (Jeong et al., 2023).
Given that the genetic compositions of mother plants and nucellar progenies are identical, nucellar embryony has been advantageous in maintaining desirable traits of rootstock or scion. However, it has posed a challenge with regard to generating genetic diversity in Citrus breeding programs. Some Citrus species, including yuzu, are known to produce seeds containing both nucellar and zygotic embryos (Zhang et al., 2018). However, it is almost impossible to distinguish zygotic seedlings from nucellar offspring by means of a visual examination. To introduce genetic diversity through self-fertilization or inter-specific hybridization, zygotic offspring should be identified within the progeny populations, which typically consist of a small number of zygotic seedlings and a larger number of nucellar seedlings.
A variety of molecular markers, such as isozyme (Soost and Williams, 1980), randomly amplified polymorphic DNA (RAPD; Yun et al., 2007; Mondal and Saha, 2014; Jin et al., 2015), simple sequence repeat (SSR; Ruiz et al., 2000; Woo et al., 2019; Singh et al., 2020), and inter-simple sequence repeat markers (ISSR; Krueger and Roose, 2003; Golein et al., 2011; Kashyap et al., 2018) have been used to identify zygotic offspring from progenies derived from self-fertilization or inter-specific hybridization in Citrus species. However, no such attempt to identify zygotic seedlings has been performed in yuzu. In this study, high-resolution melting (HRM) markers were used to identify zygotic yuzu seedlings, and an optimal set of HRM markers was determined based on the linkage relationships and segregation distortions of these HRM markers.
Materials and Methods
Plant materials and germination of yuzu seeds
Yuzu fruits harvested from three plants of the ‘Namhae 1-ho’ variety were used in this study. These yuzu plants were cultivated at the Jeollanam-do Agricultural Research and Extension Service, Wando-gun, Jeollanam-do, Republic of Korea. In earlier work (Yun et al., 2024), it was found that the genotypes of 34 HRM markers were identical in all three yuzu plants. For germination, seeds extracted from the fruits were immediately soaked in water, and the fruit flesh was completely removed by hand. The clean seeds were immersed in water for three days, and floating seeds were discarded. After a small portion of the hilum was cut with fingernail clippers, the seed coats were removed by hand. The seeds were then planted in 50-hole seed germination trays (54 × 28 cm) filled with commercial nursery bed soil. As yuzu seeds are classified as recalcitrant seeds, they were kept moist throughout all stages, i.e., from extraction to germination.
DNA extraction and HRM marker analyses
Total genomic DNA was extracted from the leaf tissues of four- to six-leaf stage seedlings using the CTAB method (Doyle and Doyle, 1987). In total, 21 HRM markers developed in previous studies (Jeong et al., 2022, 2023) were used in this study. For genotyping of the HRM markers, HRM analyses were conducted in 20 µL reaction mixtures containing 0.05 µg of a template along with 2.0 µL 10x PCR buffer, 1.0 µL forward primer (10 µM), 1.0 µL reverse primer (10 µM), 1.0 µL dNTPs (10 mM each), 0.25 U Taq polymerase (Prime Tag DNA polymerase; GeNet Bio, Nonsan, Republic of Korea), and 1.0 µL 100-fold diluted SYTO®9 green fluorescent nucleic acid stain (Thermo Fisher Scientific, Waltham, MA, USA). The primer sequences of the HRM markers are shown in Suppl. Table S1.
The PCR amplification protocol consisted of an initial denaturation step of incubation at 95°C for 10 min, followed by 45 cycles of incubation at 95°C for 10 s, 60°C for 5 s, and 72°C for 5 s. The PCR products were then heated to 95°C at a rate of 4.4°C/s, cooled to 40°C at a rate of 2.2°C/s, and heated again to 65°C at a rate of 2.2°C/s. HRM peaks were obtained using a LightCycler® 96 system (Roche Molecular Systems, Pleasanton, CA, USA) by melting the samples from 65°C to 97°C at a rate of 0.07°C/s. For genotyping of the CitRWP-MK2H marker, an HRM assay with an unlabeled probe as developed by Montgomery et al. (2007) was used. The unlabeled probe (5’-TTG-GAT-TAA-TGA-TCA-ATT-TTC-TGC-3’) with 3’ phosphorylation was utilized. A more detailed protocol is described in the literature (Jeong et al., 2023).
Constructing a linkage map of HRM markers and identifying sweet orange orthologs
Genotyping data from 21 HRM markers of 197 zygotic seedlings were used to construct a linkage map using JoinMap 4.1 (Van Ooijen and Voorrips, 2001). The linkage groups were visualized using MapChart 2.3 (Voorrips, 2002). To identify sweet orange genes showing homology with the yuzu cDNAs, which were used to develop the HRM markers, a BLAST search provided by the Citrus Genome Database (www.citrusgenomedb.org) was conducted. The target databases in the search were the sweet orange transcriptome (Citrus sinensis DHSO v3.0 genome transcripts) and the whole genome of sweet orange (Citrus sinensis DHSO v3.0 genome scaffolds).
Results
Identification of yuzu seedlings originating from zygotic embryos using molecular markers
To assess this frequency and select desirable seedlings from the zygotic embryos, a total of 837 yuzu seeds were germinated. Out of these, seedlings emerged from 707 seeds, resulting in a seed germination rate of 84.5%. Multiple seedlings were observed from 244 germinated seeds (Suppl. Fig. S1). Because seedlings from zygotic embryos were visually indistinguishable from those derived from nucellar embryos, molecular markers were used to identify the zygotic seedlings resulting from self-fertilization.
In our previous study (Jeong et al., 2022), 44 HRM markers were developed in yuzu based on SNPs in the cDNA sequences that were de novo assembled using RNA-Seq reads. Among them, ten HRM markers showing clear HRM peak patterns and heterozygous genotypes in the three mother plants of all seedlings were subjected to an analysis (Suppl. Table S1 and Fig. 1). A total of 973 seedlings were analyzed using these ten HRM markers. The results showed that at least one HRM marker exhibited homozygous genotypes in 197 seedlings, while the rest showed all heterozygous genotypes for the ten HRM markers, like the mother plants. This indicates that at least 20.2% of the seedlings originated from zygotic embryos (Suppl. Table S2). Among the 197 zygotic seedlings, 34 individuals were identified from multiple seedlings germinated from single seeds.
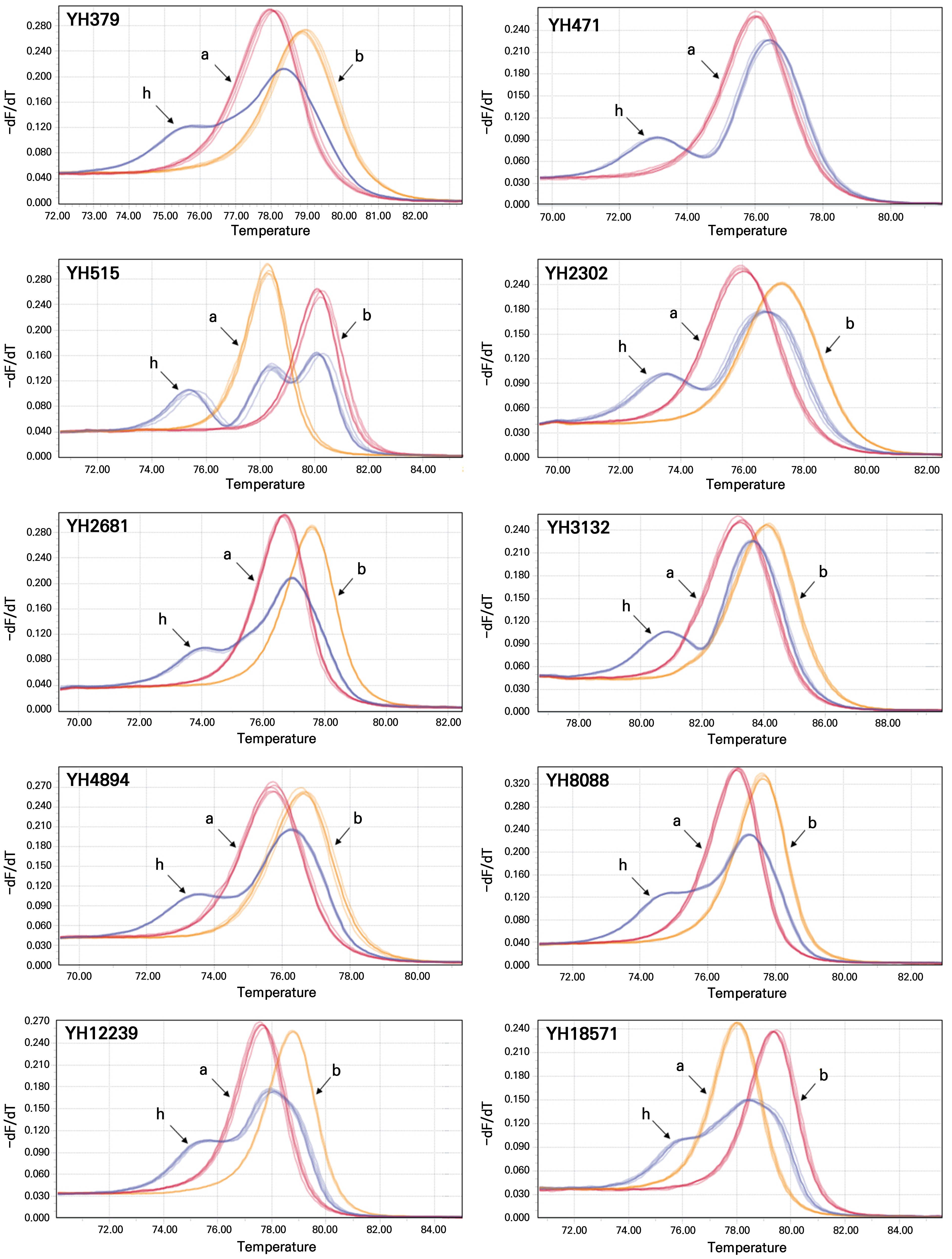
Fig. 1.
HRM peak patterns of 10 HRM markers initially used to identify zygotic seedlings. Genotypes: a and b, homozygous genotypes of control samples; h, heterozygous genotypes of three mother plants. Homozygous genotypes (a and b) are arbitrarily assigned to the HRM peaks containing lower and higher melting temperatures, respectively.
While a maximum of eight homozygous genotypes were observed in four seedlings, only one homozygous genotype was detected in seven seedlings (Suppl. Table S2). To confirm the zygotic origins further and to select the optimal set of HRM markers, all 197 zygotic seedlings were analyzed using eleven additional HRM markers. Genotypes of these eleven markers were also heterozygous in the mother plants. The results showed that at least three homozygous genotypes were identified in all 197 seedlings (Suppl. Table S3), indicating that these seedlings had derived from zygotic embryos. On average, 9.72 homozygous genotypes were observed in zygotic seedlings analyzed using 21 HRM markers here (Fig. 2A), slightly lower than the expected number of 10.5 in normal self-fertilized populations.
Regarding the segregation ratios of the 21 HRM markers, genotype ratios of nine markers were consistent with the expected 1:2:1 ratio. However, twelve other markers showed significant amounts of segregation distortion (Table 1). Among them, eight markers exhibited severe segregation distortion (p < 0.01). Interestingly, extreme levels of segregation distortion were observed in two markers (YH12239 and YH471). For YH12239, only three ‘b’ genotypes were observed, and the proportion of homozygous genotypes was slightly lower compared to most other markers (Suppl. Table S3 and Fig. 2B). With regard to the YH471 marker, the ‘b’ genotype was completely absent. In addition, the proportion of homozygous genotypes was halved in the YH471 marker (Suppl. Table S3 and Fig. 2B), indicating that seedlings containing the ‘b’ genotype of the YH471 marker did not survive.
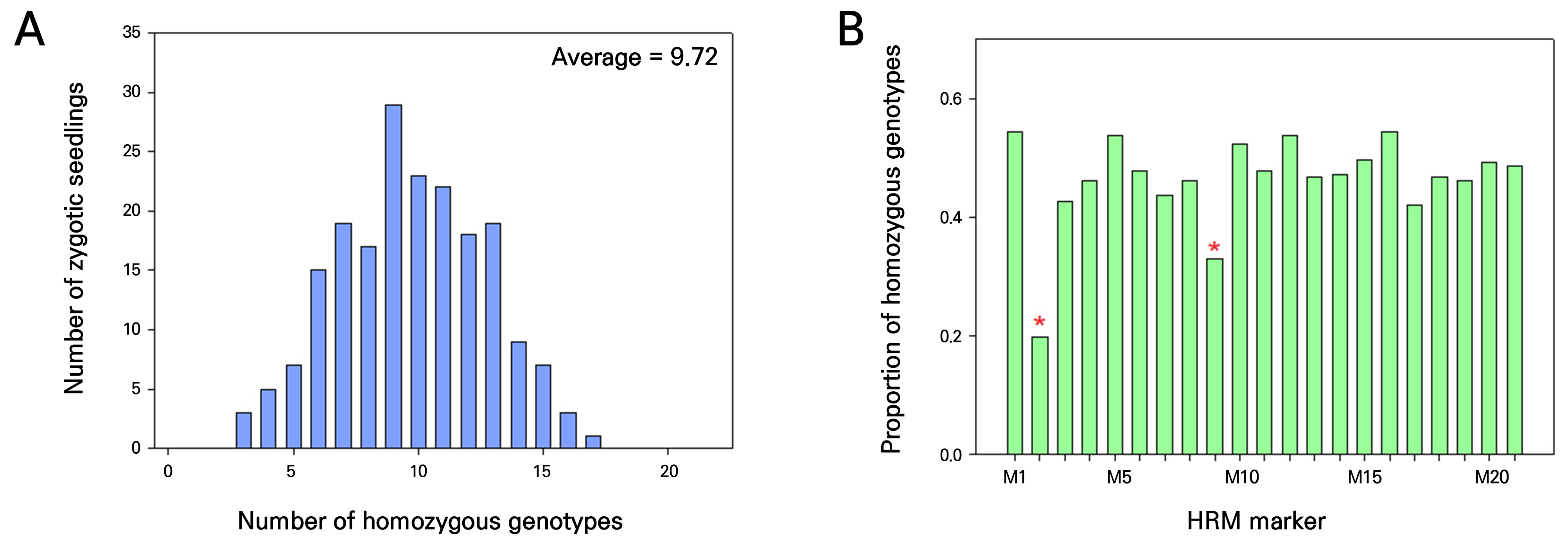
Fig. 2.
Frequencies of homozygous genotypes of 21 HRM markers in 197 zygotic seedlings. A. Histogram showing the distribution of the number of HRM markers exhibiting homozygous genotypes in individual zygotic seedlings. B. Proportion of homozygous genotypes among the 197 zygotic seedlings for each of the 21 HRM markers. M1–21, serial numbers of the 21 HRM markers. Detailed information on the HRM markers is provided in Supplementary Table 1. The YH471 and YH12239 markers showing severe segregation distortions are marked with red asterisks.
Genotyping of the CitRWP gene controlling nucellar embryony and determination of the optimal set of HRM markers based on linkage relationships
Using molecular markers developed in our previous study, yuzu was found to have a heterozygous genotype for the CitRWP gene (Jeong et al., 2023). The CitRWP genotypes of 197 zygotic seedlings were analyzed using the previously developed CitRWP-MK2H marker (Suppl. Fig. S2). This HRM marker was designed based on the mutant allele-specific SNP (Jeong et al., 2023). The segregation ratio of CitRWP genotypes did not fit the expected 1:2:1 ratio (Table 1), showing moderate segregation distortion. Significantly fewer (33) homozygous dominant genotypes conferring nucellar embryony were detected compared to the homozygous recessive wild-type genotypes (61) (Table 1).
Table 1.
Segregation ratios of genotypes for 21 HRM markers in 197 zygotic seedlings. P values lower than 0.05 are highlighted in bold. Genotypes: a, homozygous genotypes showing HRM peaks with lower melting temperatures; h, heterozygous genotypes; b, homozygous genotypes showing HRM peaks with higher melting temperatures
In total, 21 HRM markers including CitRWP-MK2H were analyzed in this study to identify the zygotic seedlings. However, the chromosomal positions of these markers were not resolved in yuzu. If any of these markers were closely linked to each other, the efficiency of identifying zygotic seedlings would be significantly reduced. To identify any close linkages of markers, the linkage relationships of all 21 markers were analyzed. Mapping all 21 markers using 197 zygotic seedlings resulted in nine independent groups at the threshold level of LOD 2.0. Five linkage groups containing at least two linked markers were produced. For example, YH784 and YH379 showed the tightest linkage with a distance of 0.8 cM (Fig. 3).
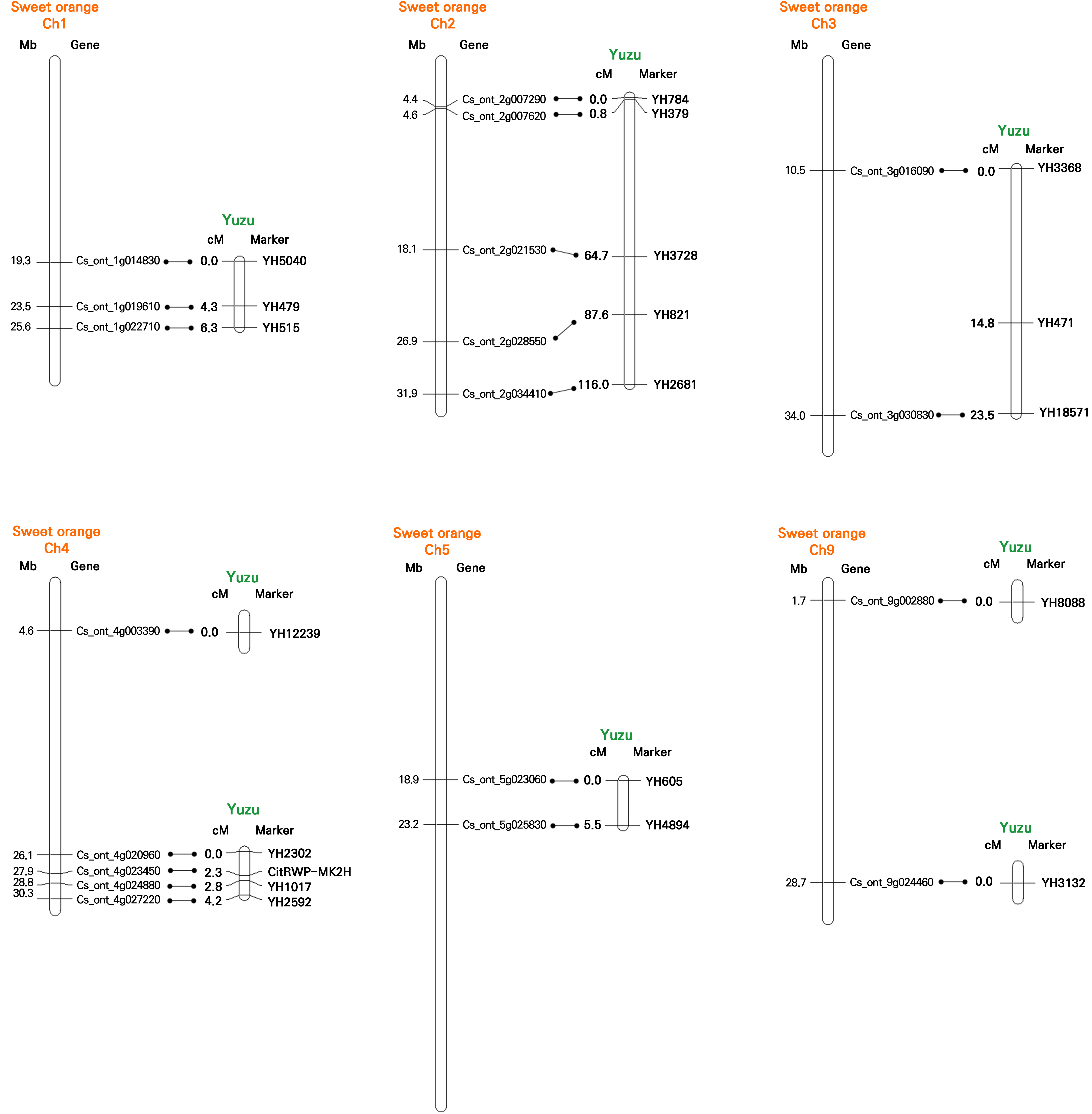
Fig. 3.
Linkage relationships of 21 HRM markers and identification of sweet orange orthologs. Left side of each panel: sweet orange physical maps. Right side of each panel: yuzu linkage maps. Homologous genes are connected with horizontal lines. Detailed information is provided in Supplementary Table 4.
Furthermore, the putative chromosomal positions of the 21 HRM markers were assessed by identifying the chromosomal positions of sweet orange orthologs showing homology with yuzu cDNAs (Suppl. Table S4). Collinearity between the yuzu and sweet orange genes was well conserved (Fig. 3). The positions of 17 linked HRM markers were perfectly collinear with those of the sweet orange homologs, except for YH471; YH471 was linked to YH3368 and YH18571 in the yuzu linkage map, but no sweet orange homolog was identified in the corresponding region of the sweet orange genome. Rather, the chromosomal region with the highest degree of homology was identified on a different chromosome, although the homology level (94.5%) was relatively low compared to other markers (Suppl. Table S4).
Considering the linkage relationships among the 21 HRM markers, the optimal set of HRM markers was determined for the efficient selection of yuzu zygotic seedlings. First, one marker with no segregation distortion was chosen from each of the five linkage groups. The CitRWP-MK2H marker was included for genotyping the CitRWP gene, despite showing moderate segregation distortion. Second, YH3132, which was not part of any linkage groups and showed no segregation distortion, was included. An analysis of 197 zygotic seedlings in this study revealed that all could be identified using the combination of these six HRM markers (CitRWP-MK2H, YH605, YH784, YH3132, YH5040, and YH18571).
Discussion
Identification of yuzu seedlings originating from zygotic embryos using molecular markers, as well as the morphological and genetic features of zygotic seedlings
Yuzu is primarily reproduced through nucellar embryony, but zygotic embryos are also produced in yuzu (Zhang et al., 2018). However, the frequency of zygotic embryo production in yuzu is still unknown. In this study, yuzu seedlings derived from zygotic embryos were successfully identified using HRM markers and the frequency of yuzu zygotic embryogenesis was assessed for the first time. Various genetic markers, including morphological, isozyme, RAPD, and SSR markers, have been used to detect zygotic seedlings in Citrus species (Kishore and Rani, 2013). We used higher-throughput SNP-based HRM markers to analyze a large number of samples in this study. SNP markers have emerged as the primary genotyping tools in plant breeding due to their suitability for automation and high-throughput analysis (Thomson, 2014). Nevertheless, only a few studies have utilized SNP-based markers for this purpose.
In this study, the proportion of yuzu seedlings with zygotic origins was estimated to be 20.2%. The frequencies of zygotic seedlings analyzed in other Citrus species vary significantly depending on the species and variety, ranging from 0.8% to more than 50% (Ashari et al., 1988; Andrade-Rodríguez et al., 2004; Golein et al., 2011; Kashyap et al., 2018). However, the numbers of samples analyzed in other studies typically amount to less than 250. In contrast, 973 yuzu seedlings were analyzed in this study. Therefore, the reliability of the frequency of zygotic seedlings obtained in this study may be relatively high relative to that of other studies. However, it is assumed that the frequency of zygotic embryogenesis in yuzu is underestimated, as a substantial number of zygotic embryos may not grow into seedlings. Generally, the growth of zygotic seedlings identified in this study was inferior to that of nucellar seedlings (Suppl. Fig. S3).
Poor growth of zygotic yuzu seedlings is attributable to inbreeding depression, which is caused by either increased homozygosity of loci with deleterious alleles or decreased heterozygosity of loci involved in hybrid vigor (Charlesworth and Charlesworth, 1999; Charlesworth and Willis, 2009; Hedrick and Garcia-Dorado, 2016). Previous studies (Swingle and Reece, 1967; Handa et al., 1986; Demarcq et al., 2021; Wu et al., 2021; Jeong et al., 2022) have suggested that yuzu is a natural hybrid between wild mandarin and Ichang papeda, although the timing of this hybridization remains unknown. Based on the observation of inferior growth in most zygotic seedlings in this study, it is assumed that natural hybridization likely occurred in ancient times, resulting in the accumulation of many deleterious mutations. Notably, twelve out of 21 HRM markers analyzed in this study showed segregation distortion in the zygotic seedling population (Table 1), possibly due to the presence of deleterious alleles. In the case of YH471, one of the two alternate homozygous genotypes was completely absent, indicating the presence of lethal alleles in the yuzu chromosomal region associated with YH471.
Despite the presence of many deleterious alleles, nucellar seedlings that maintained hybridity could have survived, as the majority of loss-of-function mutations are inherited as recessive mutations. On the other hand, zygotic seedlings that may contain homozygous genotypes of deleterious alleles may be less apt to prevail. Consequently, the genetic diversity of yuzu has become very limited. In fact, we previously analyzed the genetic diversity of 265 yuzu plants grown in Korea using SNPs identified through RNA-Seq and whole-genome resequencing (Yun et al., 2024). However, we could not find any yuzu plants derived from zygotic embryos. Therefore, marker-assisted selection of zygotic seedlings produced through self-fertilization or inter-specific hybridization is necessary to create more genetic diversity and develop new varieties of yuzu.
Development of an optimal set of HRM markers for selecting zygotic seedlings and their application in yuzu breeding
The linkage relationships of 21 HRM markers were examined in this study to identify closely linked markers. Despite using only 21 markers to construct a linkage map, this map represents the first linkage map of yuzu to the best of our knowledge. With a total of 197 zygotic seedlings used for map construction, the calculated genetic distances are likely reliable. Furthermore, the collinearity between yuzu markers and sweet orange orthologs was well conserved, except for YH471 (Fig. 3). The lack of an orthologous gene in the sweet orange genome for the YH471 marker suggests that the gene tagged by YH471 may be specific to yuzu. The observed collinearity between the two species further supports the reliability of the linkage relationships of yuzu markers. Although the whole-genome sequence of yuzu is currently unavailable, it is assumed that the synteny between yuzu and sweet orange genomes is well conserved. In earlier work (Yun et al., 2024), over 80% and 72% of RNA-Seq and whole-genome resequencing reads of yuzu, respectively, were accurately mapped to the sweet orange reference transcriptome and whole-genome sequence.
Five linkage groups, each consisting of more than two linked markers, were identified (Fig. 3). The YH784 and YH379 markers were found to be linked to each other with a distance of 0.8 cM. Only three zygotic seedlings were identified as recombinants between these two markers showing different genotypes (Suppl. Table S3). Therefore, analyzing these two markers for the identification of zygotic seedlings may be redundant. Likewise, when other markers within the same linkage groups are used together, the efficiency of zygotic seedling selection may be diminished. However, most studies focusing on the marker-assisted selection of zygotic seedlings have not revealed any linkage relationships among markers. Considering the positions of HRM markers within the linkage groups, an optimal set of six HRM markers was selected in this study. This set of markers will be efficiently used to identify zygotic seedlings in yuzu breeding programs in the future.
The CitRWP-MK2H marker used to genotype the CitRWP gene controlling nucellar embryony was included in the optimal set of markers. This marker, developed based on the mutant-specific SNP (Jeong et al., 2023), allowed for an accurate prediction of CitRWP gene genotypes. Zygotic seedlings containing the homozygous genotype of the wild-type CitRWP allele may not exhibit the trait of nucellar embryony, resulting in offspring derived from zygotic embryos. These yuzu offspring can contribute to increased genetic diversity in yuzu breeding programs. While zygotic seedlings typically show poor growth, some have exhibited vigor comparable to nucellar seedlings (Suppl. Fig. S3). Therefore, the optimal set of HRM markers developed in this study will be valuable as they can ensure the effective selection of a limited number of desirable and promising zygotic seedlings.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20240056_Fig_S1.jpg
Supplementary Fig. S1. Yuzu seedlings
- HORT_20240056_Fig_S2.jpg
Supplementary Fig. S2. CitRWP marker analysis
- HORT_20240056_Fig_S3.jpg
Supplementary Fig. S3. nucellar vs zygotic
- HORT_20240056_Table_S1.xlsx
Supplementary Table S1. Primer sequences of HRM markers used in this study
- HORT_20240056_Table_S2.xlsx
Supplementary Table S2. Genotypes of 973 yuzu seedlings for the 10 HRM markers. The homozgyous genotypes (a and b) are arbitarily assinged for low and high melting peaks, respectively
- HORT_20240056_Table_S3.xlsx
Supplementary Table S3. Genotypes of 21 HRM markers of the 197 seedlings originating from zygotic embryogenesis
- HORT_20240056_Table_S4.xlsx
Supplementary Table S4. List of sweet orange genes showing the highest homology with the yuzu cDNA containing the SNPs used to design 21 HRM markers


