Introduction
Materials and Methods
Plant Materials
Sequence Analysis of Hypervariable Regions in Chloroplast Genomes
RNA-seq Analysis and Identification of Polymorphic Sequences in the Transcriptome
Construction of Phylogenetic Trees and Identification of Yuzu Homologs of 103 Single-copy Genes Conserved Among Citrinae Species
Genotyping of SNPs in Yuzu Single-copy Gene Homologs and HRM Markers
Results
Identification of a Putative Maternal Species of Yuzu using Hypervariable Regions in Chloroplast Genome Sequences of Citrus Species
Identification of a Putative Paternal Species of Yuzu using Polymorphism in Nuclear Single-copy Genes of Citrus Species
Discussion
Identification of Putative Parental Species of Yuzu by Sequence Analyses of Chloroplast Genome and Nuclear Single-copy Genes
Implication of a Hybrid Nature of Yuzu in Establishment of an Optimal Strategy for Yuzu Breeding
Introduction
Yuzu (Citrus junos Sieb. Ex Tanaka), which is a Citrus species, is known to have originated from China. It is widely grown in Korea and Japan (Swingle and Reece, 1967). Thanks to the pleasant fragrance of its fruits, the demand and production of yuzu have sharply increased in Korea since 2000 (Kim et al., 2015). Although yuzu is rarely consumed as fresh fruits due to its tart flavor, it has been used as an important ingredient in food products and cuisines. In addition, yuzu is commercially processed as cosmetics, perfumes, and oils for aromatherapy (Matsumoto et al., 2016). Yuzu is also known to possess several health-promoting activities such as antioxidant, anti-platelet, anti-inflammatory, and antidiabetic effects (Hirota et al., 2010; Yu et al., 2011; Kim et al., 2013; Lee et al., 2020).
Although genomic analyses of other major Citrus species such as sweet orange and mandarins have been extensively performed (Gmitter Jr. et al., 2012; Omura and Shimada, 2016), few genetics and genomics studies on yuzu have been carried out. So far, whole genome sequences of nine Citrinae species, including wild and cultivated accessions, have been reported (Rao et al., 2021). Among the Citrus species, sweet orange (C. sinensis) was the first to have its genome de novo assembled. Comparative analysis with other citrus genomes has suggested that sweet orange might have originated from a backcross hybrid between pummelo and mandarin (Xu et al., 2013). Sweet orange is commercially the most important citrus. Its worldwide production is ranked first among Citrus species (http://faostat.fao.org).
There is a general agreement that citrus can be classified into three basic species: mandarin (C. reticulata), pummelo (C. maxima), and citron (C. medica). Most cultivars might have derived from these basic species and their hybrids (Rao et al., 2021). In addition, papeda (C. micrantha) has also been considered as a basic species (Curk et al., 2016). Since the first report of a draft genome sequence of satsuma mandarin (Shimizu et al., 2016), genomes of diverse wild mandarins have been sequenced and their detailed domestication history has been elucidated (Wang et al., 2017). Draft genome sequences of two other basic species of pummelo and citron and two wild species of Ichang papeda (C. cavaleriei) and Chinese box-orange (Atalantia buxifolia) have been assembled and phylogeny analyses of basic and wild species have been performed using 103 single-copy genes conserved among these species (Wang et al., 2017).
Regarding organelle genomes of Citrus species, a 640-kb complete mitochondrial genome sequence containing 63 genes of sweet orange has been reported (Yu et al., 2018). The complete chloroplast genome sequence has also been assembled for sweet orange (Bausher et al., 2006). Later on, complete chloroplast genome sequences of 34 Citrus species have been assembled by mapping raw sequences to sweet orange sequences as a reference, and five hypervariable regions on citrus chloroplast genomes have been identified (Carbonell-Caballero et al., 2015). Recently, the complete chloroplast genome sequence of Ichang papeda has been reported (Zhang et al., 2020). Swingle and Reece (1967) have proposed that Ichang papeda is a seed parent of yuzu.
However, the origin of yuzu remains controversial. Swingle and Reece (1967) have suggested that yuzu is a natural hybrid between Ichang papeda and mandarin. Handa et al. (1986) have also proposed that yuzu originates from a hybrid. A recent study (Demarcq et al., 2021) has shown that yuzu might be a hybrid between Ichang papeda and mandarin based on analysis of ancestral species diagnostic single nucleotide polymorphism (SNP) markers. However, Tanaka (1954) and Tanaka and Taninak (1960) stated that yuzu is not a hybrid and assigned it a scientific name, C. junos. Hirai et al. (1986) and Uzun et al. (2009) suggested that yuzu is not closely related to Ichang papeda or mandarins based on analyses of isozymes and sequence-related amplified polymorphism (SRAP) markers, respectively. More recently, Shimizu et al. (2016) have shown that yuzu contains a unique cytotype and refuted the hypothesis of its hybrid origin based on analysis of 123 DNA markers.
In this study, we inferred the origin of yuzu based on extensive nucleotide sequence analyses of nuclear and chloroplast genomes. A putative maternal parent of yuzu was identified using sequence analysis of hypervariable regions of citrus chloroplast genomesidentified byCarbonell-Caballero et al. (2015). A putative paternal parent and the hybrid origin of yuzu were proposed on the basis of sequence analysis of 80 nuclear single-copy genes identified from de novo assembled transcriptome of yuzu.
Materials and Methods
Plant Materials
A total of 37 accessions, including papeda relatives, Ichang papeda-like citrus, and yuzu, were used in the study (Suppl. Table 1). Eight accessions of papeda relatives and one Ichang papeda-like citrus were used to analyze their phylogenetic relationships with yuzu based on variations of chloroplast genome sequences. The origin of Ichang papeda-like citrus was unclear. Four yuzu accessions sampled from a collection of citrus germplasm at Jeju Agricultural Research and Extension Service were used to assess variations in chloroplast genome sequences among yuzu accessions. Two yuzu cultivars, ‘Dajungeum’ (or ‘Tadanishiki’) and ‘Harim12’, were used for RNA-sequencing (RNA-seq) analysis (Suppl. Fig. 1). Twenty-two yuzu samples collected from nine orchards in Goheung province, Korea were used to analyze high resolution melting (HRM) markers (Suppl. Tables 1 and 2).
Sequence Analysis of Hypervariable Regions in Chloroplast Genomes
Five hypervariable regions spanning 18,097 bp in Citrus chloroplast genomes were sequenced from three yuzu cultivars ‘Dajungeum’, ‘Harim12’, and ‘Yamane’ (Table 1). These regions have been previously identified by comparative analysis of 34 complete citrus chloroplast genome sequences (Carbonell-Caballero et al., 2015). To amplify these regions, primer pairs were designed based on the sequence of the sweet orange chloroplast genome (Genbank accession number: DQ864733). Primer sequences are shown in Suppl. Table 3. For identification of papeda relatives closely related to yuzu, sequences of only one hypervariable region showing the highest variation were obtained from nine papeda relatives. Total genomic DNA was extracted from leaf tissues of yuzu using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987).
Table 1.
Hypervariable regions in citrus chloroplast genomes analyzed in this study
PCR amplification was performed in a 25-µL reaction mixture containing 0.1 µg template, 2.5 µL 10× PCR buffer, 0.5 µL forward primer (10 µM), 0.5 µL reverse primer (10 µM), 0.5 µL dNTPs (10 mM each), and 0.25 µL polymerase mix (Advantage 2 Polymerase Mix, Takara Bio, Shiga, Japan). PCR amplification consisted of an initial denaturation step at 95°C for 4 min; 10 cycles of 95°C for 30 s, 65°C (0.8°C decrements in each cycle) for 30 s, and 72°C for 1 min; 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min; and a final 10-min extension step at 72°C. After confirming successful PCR amplification by visualizing PCR products on 1.5% agarose gels after ethidium bromide staining, PCR products were purified for sequencing using a QIAquick PCR Purification kit (QIAGEN, Valencia, CA, USA). Sequencing was performed by Macrogen (Seongnam, Republic of Korea). Any gaps in long or complicated sequences were filled by primer walking.
RNA-seq Analysis and Identification of Polymorphic Sequences in the Transcriptome
To perform RNA-seq, total RNA was extracted from two pooled samples of young leaves and floral buds of ‘Dajungeum’ and ‘Harim12’, respectively, using an RNA extraction kit (RNeasy Plant Mini Kit, QIAGEN) following the manufacturer’s instructions. RNA-seq analysis was carried out by Phyzen Genomics Institute (Seoul, Republic of Korea). Details about RNA quality and library construction are shown in Suppl. Table 4. Transcriptome sequences were produced using a HiSeq X Ten platform (Illumina, Hayward, CA, USA). Transcriptome sequences of Dajungeum and Harim12 were deposited into the Sequence Read Archive (SRA) database under accession numbers SRR16883166 and SRR16883167, respectively. After trimming raw reads using Trimmomatic software (Bolger et al., 2014), trimmed reads were de novo assembled into reference contigs using Trinity software (Haas et al., 2013). Trimmed reads of two cultivars were separately mapped to the yuzu reference transcriptome using BWA software (Li and Durbin, 2009). SNP discovery was carried out using GATK (Van der Auwera and O’Connor, 2020). Versions and options of the software used are shown in Suppl. Table 5.
Construction of Phylogenetic Trees and Identification of Yuzu Homologs of 103 Single-copy Genes Conserved Among Citrinae Species
Nucleotide sequences of chloroplast genome and single-copy genes of Citrus species were aligned using BioEdit software (Hall, 1999). Gaps in alignments were removed using Gblocks software (Castresana, 2000). Phylogenetic trees were constructed with MEGA version X (Kumar et al., 2018) using the neighbor-joining method. Node support of the phylogenetic tree was assessed by 1,000 bootstrap replicates. Mean pairwise genetic distances of hypervariable regions in chloroplast genomes were calculated using MEGA software.
In a previous study (Wang et al., 2017), 103 single-copy genes were identified from eight Citrinae species. Yuzu homologs of these genes were identified among contigs assembled by RNA-seq using BioEdit software (Hall, 1999). Coding sequences of other Citrinae species corresponding to yuzu homologs were identified from a coding sequence database (the Citrus Pan-genome to Breeding Database, citrus.hzau.edu.cn) for Chinese box-orange (A. buxifolia), citron (C. limon), mandarin (C. reticulata), sweet orange (C. sinensis), Ichang papeda (C. cavaleriei), and pummelo (C. grandis). A physical map of sweet orange single-copy genes of which homologs were identified in yuzu was drawn using MapChart 2.3 (Voorrips, 2002).
Genotyping of SNPs in Yuzu Single-copy Gene Homologs and HRM Markers
For genotyping of SNPs in single-copy gene homologs, conserved sequences among yuzu, citron, mandarin, Ichang papeda, and pummelo were aligned using BioEdit software (Hall, 1999). Chinese box-orange and sweet orange sequences were excluded for simplicity of analyses since Chinese box-orange is too distantly related to yuzu and sweet orange is not a basic species. Genotypes of SNPs in yuzu sequences were determined by visual examination of mapped RNA-seq reads to the reference transcriptome using IGV viewer software (Robinson et al., 2011). Genotypes of SNPs were considered heterozygous if the number and proportion of reads containing a minor allele were at least three and more than 20%, respectively (Fig. 1).
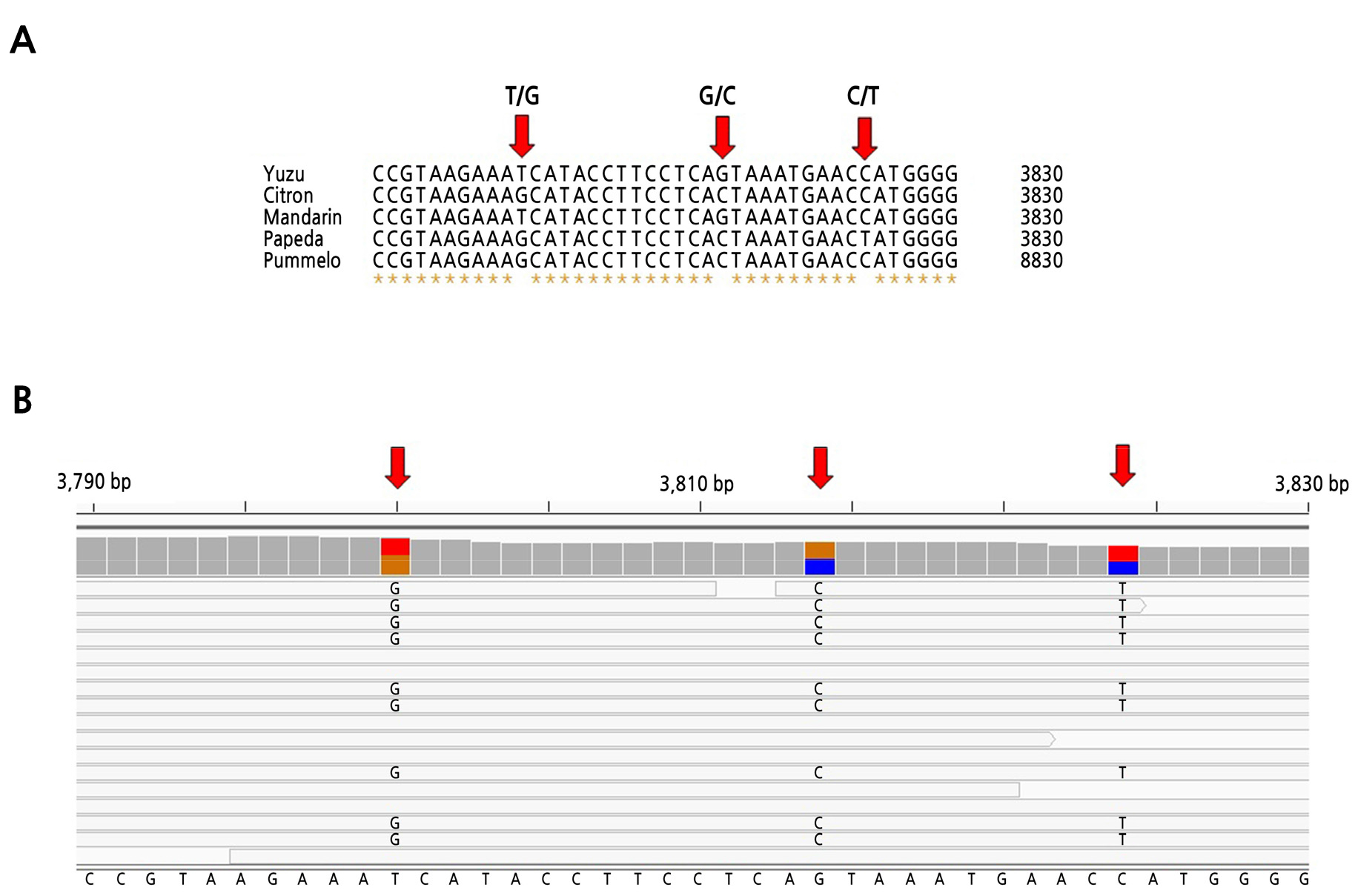
Fig. 1.
Genotyping of single nucleotide polymorphisms (SNPs) in single-copy genes of yuzu. A. Alignment of single-copy genes of five Citrus species. Positions of SNPs are indicated by vertical arrows. Genotypes of yuzu are shown above vertical arrows. B. An IGV viewer image of mapping of reads produced by RNA-seq to aligned sequences shown in Fig. 1A. Vertical arrows indicate positions of SNPs. Aligned sequences are parts of a single-copy gene (SCG84).
For genotyping of HRM markers, SYTO®9 green fluorescent nucleic acid stain (Thermo Fisher Scientific, Waltham, MA, USA) and a LightCycler® 96 system (Roche Molecular Systems, Pleasanton, CA, USA) were used. A detailed protocol was described in a previous study (Kim and Kim, 2019). Primer sequences of HRM markers developed in this study are shown in Suppl. Table 6.
Results
Identification of a Putative Maternal Species of Yuzu using Hypervariable Regions in Chloroplast Genome Sequences of Citrus Species
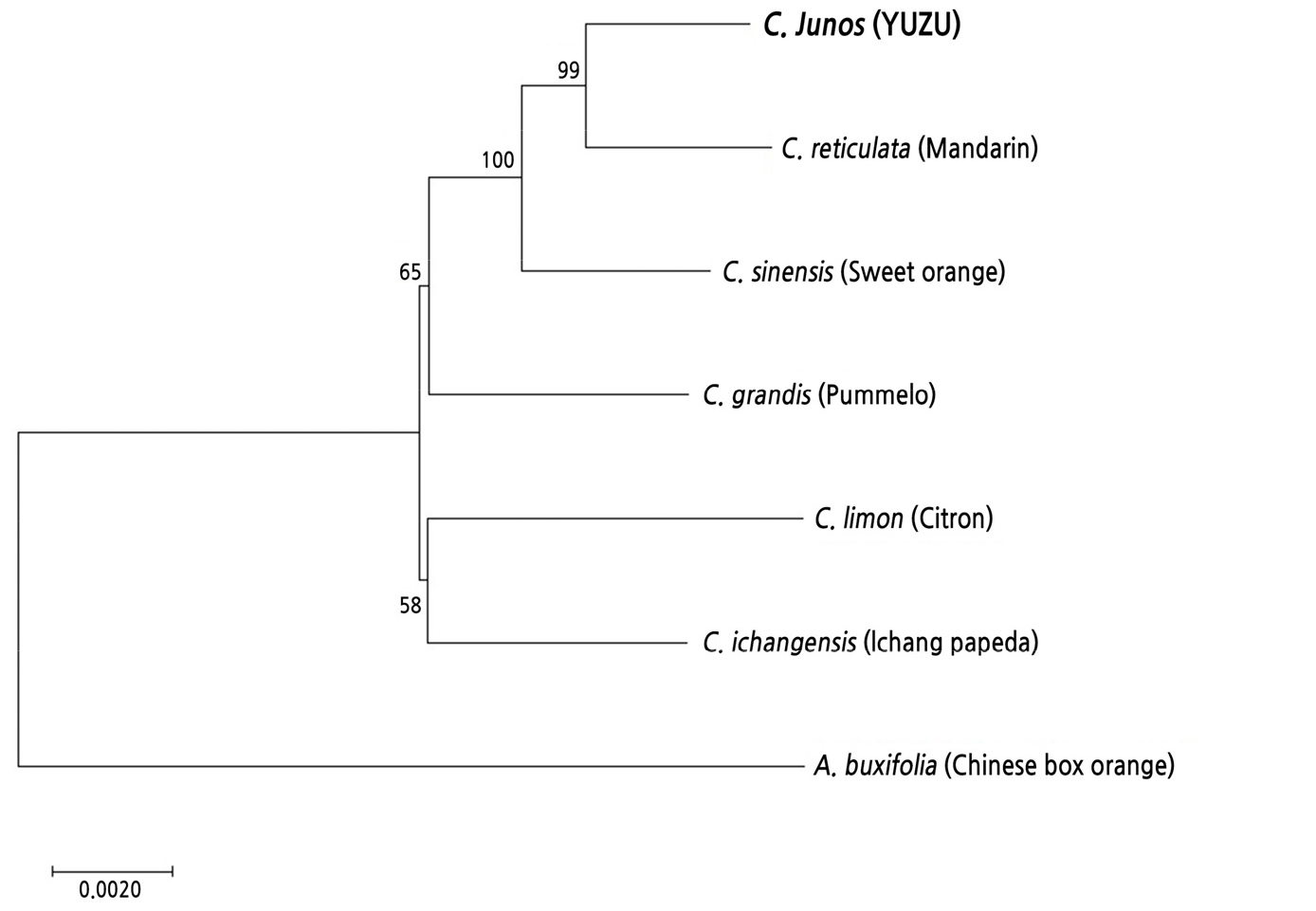
Many Citrus species are known to be natural hybrids. Thus, a putative seed parent of yuzu was inferred by analyzing variations in chloroplast genome sequences. Based on a previous report (Carbonell-Caballero et al., 2015) of five hypervariable regions in complete chloroplast genomes of 34 Citrus species, nucleotide sequences of these regions spanning 18,097 bp in the sweet orange chloroplast genome were obtained from yuzu. Chloroplast genome sequences of sweet orange (DQ864733) were used as a reference (Table 1). Combined Sequences from five regions covering 17,531 bp were assembled from yuzu. These nucleotide sequences were deposited into GenBank under accession numbers OM303015 to OM303019. Homologous sequences of these 17,531-bp regions were identified from 18 Citrus and one Atalantia species for which complete chloroplast genome sequences were publicly available. A phylogenetic tree was constructed using alignments of these homologous sequences (Fig. 2). The tree showed that yuzu was most closely related to Ichang papeda. Nucleotide sequences of yuzu and Ichang papeda shared 99.8% identities with each other.
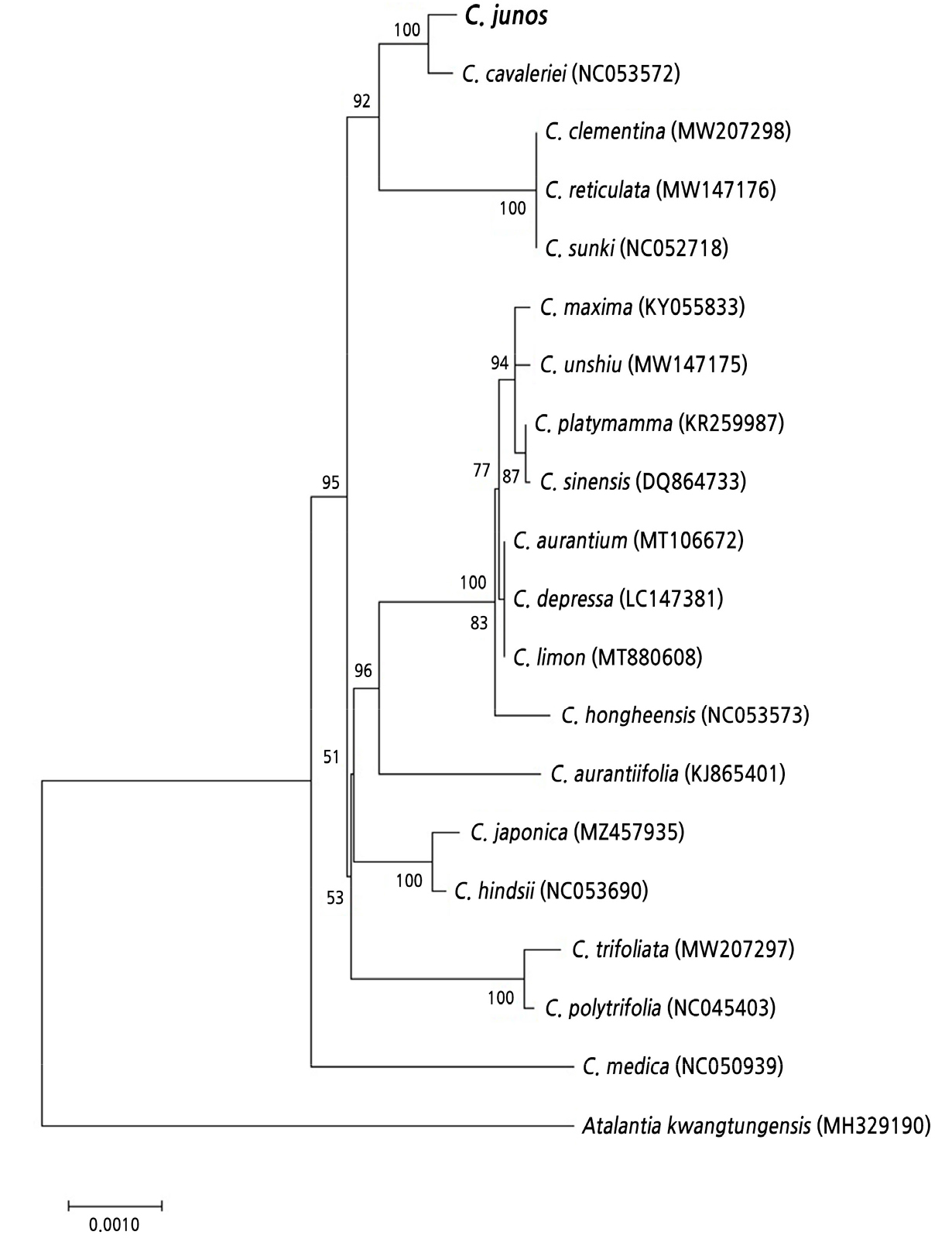
Fig. 2.
Phylogenetic tree of chloroplast genome sequences of yuzu and other Citrus species. The sequence of Atalantia kwangtungensis was used as an outgroup. Concatenated sequences of five hypervariable regions (17,531 bp) were used to construct the phylogenetic tree. GenBank accession numbers of other species are shown in parentheses.
To identify more closely related species and to assess variations among yuzu accessions, additional papeda relatives and yuzu accessions were analyzed. Among five hypervariable regions in chloroplast genomes, the Cp1 region was selected for further analyses since it had the highest variation according to mean pairwise genetic distances among 20 species (Suppl. Fig. 2). A phylogenetic tree constructed using Cp1 sequences of 10 papeda relative accessions showed that Ichang papeda was still the closest relative of yuzu (Suppl. Fig. 3). These results imply that the maternal parent of yuzu might be Ichang papeda or its close relatives. When Cp1 sequences of four yuzu accessions were analyzed, only a single 1-bp insertion/deletion (InDel) was identified in ‘Yamane’ compared with other yuzu accessions. Sequences of the other three accessions were identical to those of ‘Dajungeum’ and ‘Harim12’. The presence of the 1-bp InDel in ‘Yamane’ was confirmed by repetition of PCR product sequencing (Suppl. Fig. 4). In the case of ‘Yamane’, four other hypervariable regions were sequenced. However, no additional variation was identified.
Identification of a Putative Paternal Species of Yuzu using Polymorphism in Nuclear Single-copy Genes of Citrus Species
A total of 103 single-copy genes conserved among eight Citrinae species were identified through comparative analyses of whole genome sequences in a previous study (Wang et al., 2017). To identify yuzu homologs of these single-copy genes, transcriptome sequences of two yuzu cultivars (‘Dajungeum’ and ‘Harim12’) were obtained by RNA-seq (Suppl. Table 7). A total of 119,137 contigs were de novo assembled using trimmed reads (Suppl. Table 8). Eighty contigs showing homology with single-copy genes were identified using a local BLAST search (Suppl. Table 9). Since yuzu contigs were assembled using transcriptome sequences, coding sequences corresponding to these 80 single-copy genes were identified from coding sequence databases of Chinese box-orange, citron, mandarin, sweet orange, papeda, and pummelo, respectively (Suppl. Table 9).
Conserved regions among yuzu and six Citrinae species were identified by multiple sequence alignments. Sequences of these conserved regions were used for further analyses. A phylogenetic tree was constructed using concatenated sequences (69,309 bp) of conserved regions of 80 single-copy genes. In contrast to the tree based on chloroplast genome sequences, the phylogenetic tree based on nuclear genes showed that yuzu was most closely related to mandarin (Fig. 3). These results suggest that the pollen parent of yuzu might be a mandarin. Since yuzu was probably derived from a hybrid between Ichang papeda and mandarin, significant portions of the yuzu genome were assumed to have both mandarin and Ichang papeda origins.
To assess the heterozygosity level of the yuzu genome, SNPs in the yuzu transcriptome were identified by mapping trimmed reads to the reference transcriptome (Suppl. Table 10). More than 300,000 variants were detected in both ‘Harim12’ and ‘Dajungeum’ (Suppl. Table 11). Although most variants seemed to be derived from intra-varietal SNPs, some of them were polymorphic between ‘Harim12’ and ‘Dajungeum’. To confirm the polymorphism between these two cultivars, 44 HRM markers were developed based on SNPs or InDels. Genotypes of all HRM markers were different between the two cultivars (Suppl. Fig. 5 and Suppl. Table 2). However, 26 yuzu samples (all samples except for ‘Harim12’ and HR143) showed identical genotypes for all HRM markers (Suppl. Table 2). These results implied that ‘Harim12’ and HR143 were clonally propagated from the same cultivar and that the original cultivar might be bred by hybridization between yuzu and other Citrus species. Fruit and leaf morphologies of ‘Harim12’ were somewhat different from those of other typical yuzus (Suppl. Fig. 1), although sequences of five hypervariable regions in the chloroplast genome were identical to those of ‘Dajungeum’. Therefore, ‘Harim12’ was excluded from further analyses.
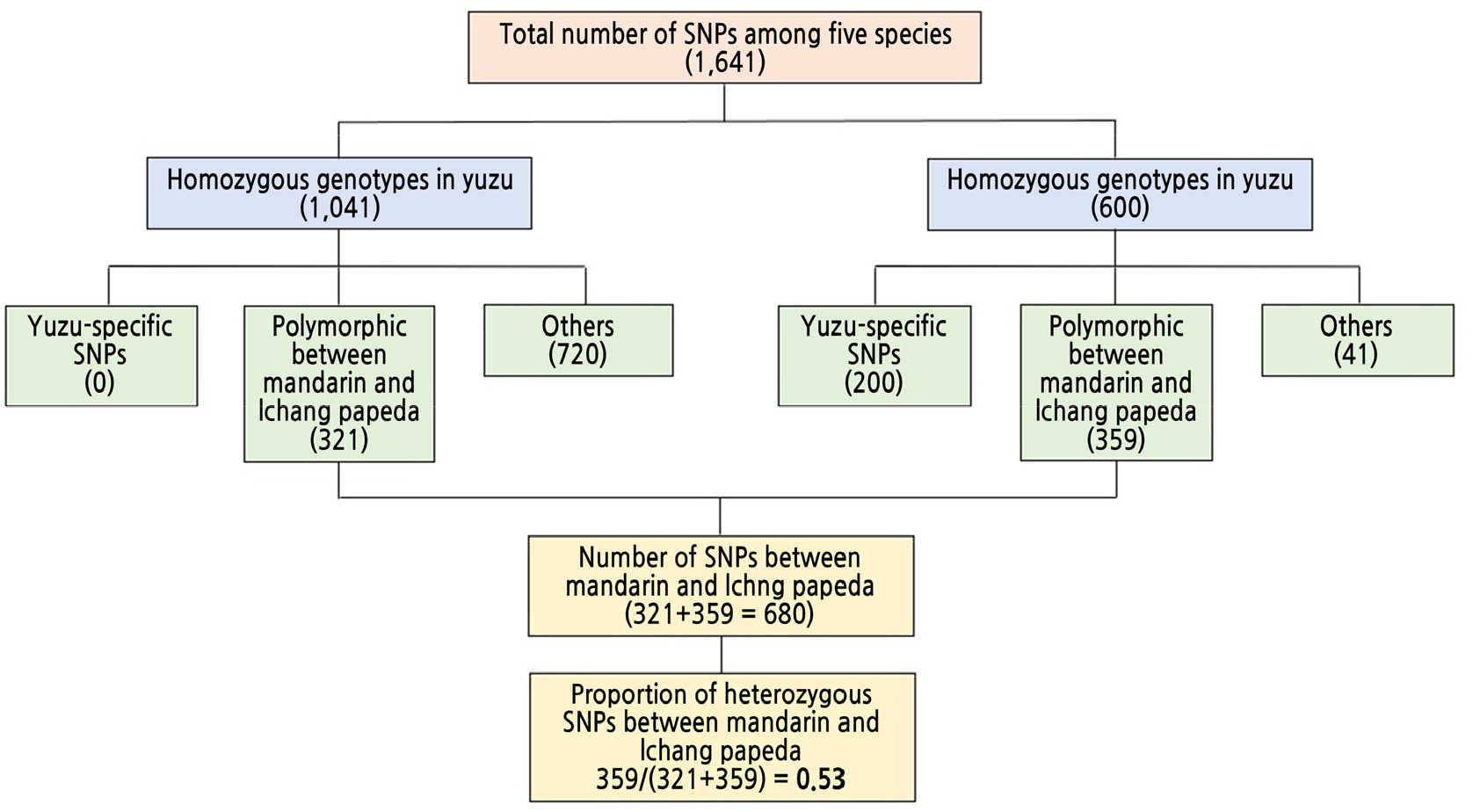
To assess the portion of hybrid origin in the yuzu genome, SNPs in the 80 single-copy genes were analyzed. In the case of heterozygous SNP loci in yuzu, contrasting SNP alleles were identified by visual examination of read mapping (Fig. 1). A total of 1,641 SNPs among five Citrus species were identified (Fig. 4). Regarding these 1,641 SNPs, yuzu’s alleles shared 89.8%, 85.7%, 77.5%, and 67.7% identities with those of mandarin, Ichang papeda, pummelo, and citron, respectively. In the case of yuzu-specific SNP alleles, no homozygous SNP allele was identified, although 200 yuzu-specific alleles were present in heterozygous SNPs (Fig. 4). Between mandarin and Ichang papeda, 321 and 359 SNPs were homozygous and heterozygous in yuzu, respectively, indicating that more than half of these SNPs might have derived from hybrids between mandarin and Ichang papeda (Fig. 4).
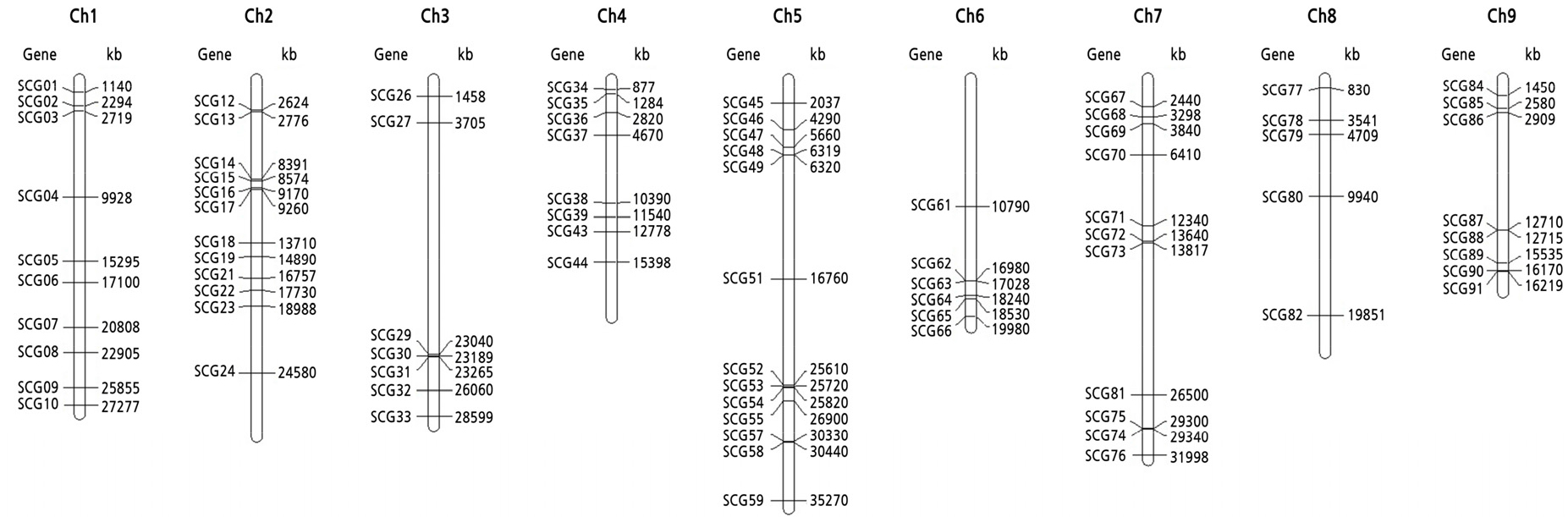
For the 80 single-copy genes, genes were considered hybrids if proportions of heterozygous SNPs consisting of mandarin and Ichang papeda alleles were higher than those of homozygous SNPs. In other cases, origins of single-copy genes were determined based on homologies with four other species. Forty-three single-copy genes were classified into hybrids. Fifteen and 11 genes were classified into mandarin and Ichang papeda origins, respectively. In addition, three genes and one gene showed the highest homologies with pummelo and citron sequences, respectively (Fig. 5, Suppl. Table 12). To investigate any clustering patterns of genes with hybrid origins, chromosomal positions of the 80 single-copy genes were identified in the sweet orange genome since synteny among Citrus species was well conserved. Eighty genes were distributed throughout nine chromosomes. Genes with hybrid origins were also distributed over all chromosomes (Fig. 6). Although the number of genes analyzed in this study was small, there were several clusters of genes with the same origins. For example, a cluster consisting of five genes with hybrid origins was observed in chromosome 1 (Fig. 6).
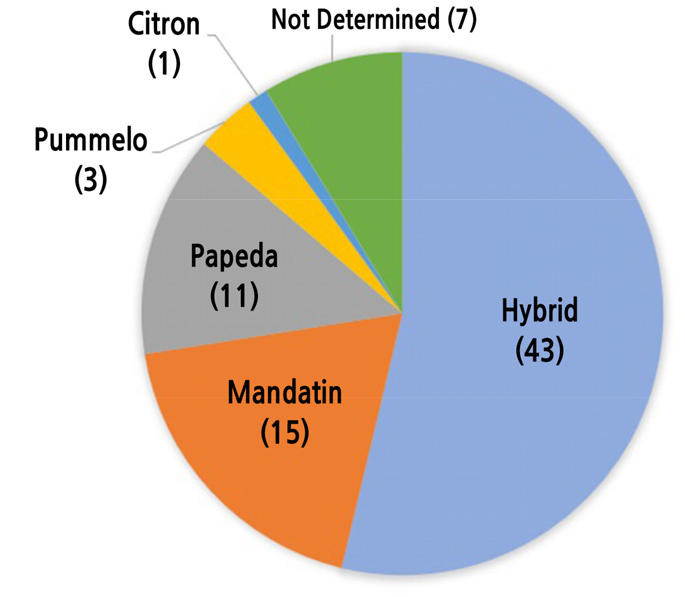
Fig. 5.
Pie chart showing proportion of putative origins of 80 yuzu single-copy genes. Detailed information is shown in Suppl. Table 12.
Discussion
Identification of Putative Parental Species of Yuzu by Sequence Analyses of Chloroplast Genome and Nuclear Single-copy Genes
Many indigenous citrus cultivars are known to originate from natural hybrids. Their parentage has been identified using molecular makers developed on the basis of organelle and nuclear genome sequences (Shimizu et al., 2016; Woo et al., 2020; Kim et al., 2021; Yi et al., 2021). In this study, we hypothesized that yuzu might have originated from natural hybridization between Ichang papeda and mandarin based on sequence analyses of chloroplast genomes and nuclear genes. Initially, Swingle and Reece (1967) considered yuzu as a hybrid between Ichang papeda and mandarin. Thus, they did not assign any scientific name to yuzu. Meanwhile, Tanaka and Taninaka (1960) classified Ichang papeda and yuzu in the same subgenus (Metacitrus) and section (Osmocitrus) but different subsections (Protosmocitrus and Euosmocitrus, respectively). They assigned the scientific name C. junos to yuzu. However, Shimizu et al. (2016) suggested that yuzu might not be an offspring of Ichang papeda because 31 out of 123 DNA markers based on nuclear genomes and four out of 11 DNA markers based on organelle genomes are not matched with each other.
Results of the present study supported the hypothesis of Swingle and Reece (1967). In the case of the seed parent, Shimizu et al. (2016) used only 11 simple sequence repeat (SSR) markers to classify cytotypes of citrus cultivars. In contrast, we used nucleotide sequences of four hypervariable regions in citrus chloroplast genomes spanning 17,531 bp. Because the reliability of parentage tests is proportional to the number of analyzed loci, sequence analysis in this study might be more reliable than that of Shimizu et al. (2016). Since Ichang papeda and yuzu shared 99.8% nucleotide sequence identities, yuzu’s seed parent was probably Ichang papeda or its close relatives. A 1-bp InDel was identified in the Cp1 region of the chloroplast genome of one yuzu accession ‘Yamane’. Although this InDel is positioned in an error-prone poly-A sequence (Suppl. Fig. 4), existence of any intra-specific variation in chloroplast genomes indicates that yuzu might have originated from an ancient event of natural hybridization, not a recent artificial hybridization. In addition, since genotypes of 44 HRM markers were identical between ‘Yamane’ and typical yuzu samples (Suppl. Table 2), it was assumed that new cultivars of yuzu were rarely produced by sexual reproduction.
Regarding the pollen parent of yuzu, mandarin was proposed as a paternal species in this study based on sequence analysis of 80 nuclear single-copy genes. It might be difficult to identify a pollen parent using DNA markers for a couple of reasons. First, genotyping errors of molecular markers might have a significant effect on the reliability of tests. Although transferability of SSR markers developed in Citrus species is relatively high compared to those of other crops (Luro et al., 2008), genotyping errors might occur when diverse species are analyzed together. Second, homologous sequences tagged by molecular markers might complicate genotyping of markers. For this reason, we selected single-copy genes conserved among Citrus species to avoid complexity derived from the presence of multiple homologs. In addition, since genotypes of SNPs were confirmed by visual examination of read mapping using IGV viewer (Fig. 1), the reliability of the SNP analysis performed in this study would be higher than that of molecular markers. In particular, heterozygous SNPs present in yuzu genes could be efficiently genotyped. Since the phylogenetic tree constructed using approximately 70-kb sequences of single-copy genes showed that yuzu was most closely related to mandarin, and many heterozygous SNPs contained both Ichang papeda and mandarin alleles, mandarin was probably the pollen parent of yuzu.
Implication of a Hybrid Nature of Yuzu in Establishment of an Optimal Strategy for Yuzu Breeding
More than half of the 80 yuzu single-copy genes analyzed in this study showed heterozygous genotypes (Fig. 5). It could be assumed that a significant portion of yuzu genomes might have remained as hybrids consisting of Ichang papeda and mandarin genomes following an initial natural hybridization event. Two factors may contribute to the high level of heterozygosity of the yuzu genome. First, yuzu is predominantly reproduced through asexual nucellar embryony, a sporophytic apomixis (Dewi et al., 2013). Nucellar embryony is widespread among most commercial cultivars of sweet orange, mandarin, grapefruit, and lemon (Xu et al., 2021). Because of nucellar embryony, the heterozygous genetic make-up of yuzu might have been maintained without meiotic recombination.
Secondly, yuzu can produce zygotic embryos through normal meiosis, although the frequency of its zygotic embryo production is very low (Dewi et al., 2013). However, the probability of producing superior progenies through self- fertilization seems to be not so high if the major traits and fitness of yuzu are significantly affected by dominance effects of heterozygous genotypes. Although yuzu cultivated in Korea are propagated by both grafting and seed germination, the genetic make-up of yuzu grown in Korea seems to be almost identical (Suppl. Table 2). This fact reflects that chance seedlings originating from zygotic embryos may not be selected due to inferior phenotypes.
However, if a large number of chance seedlings are produced, superior individuals could be selected from a large collection of variants. HRM markers developed in this study could be effectively used to identify zygotic seedlings at early stages. Since yuzu is perennial with a long juvenile period, marker-assisted selection is particularly useful for yuzu compared with other annual crops. Alternatively, superior cultivars might have been produced by artificial hybridization of papedas and mandarins. Although a more detailed analysis of the organization of the yuzu genome is required, a significant portion of single-copy genes distributed throughout all chromosomes was shown to have heterozygous genotypes containing both Ichang papeda and mandarin alleles. Sixty-eight out of 80 single-copy genes contained at least one such heterozygous SNP (Suppl. Table 12). Therefore, cross breeding between papeda and mandarin accessions might be more effective than selecting desirable progenies originating from self-fertilization. Indeed, Swingle and Reece (1967) have stated that a hybrid between Ichang papeda and satsuma mandarin shows a high similarity to yuzu. Results presented in this study provide important information to establish an optimal strategy for developing improved yuzu cultivars.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20220038_Table_1s.pdf
Supplementary Table 1. List of papeda relative and yuzu accessions used in this study
- HORT_20220038_Table_2s.pdf
Supplementary Table 2. HRM marker genotypes of yuzu samples used in this study
- HORT_20220038_Table_3s.pdf
Supplementary Table 3. List of primers used in amplification of chloroplast genome sequences
- HORT_20220038_Table_4s.pdf
Supplementary Table 4. Information about RNA extraction and library construction for RNA-Seq analysis
- HORT_20220038_Table_5s.pdf
Supplementary Table 5. Versions and options of software used in RNA-Seq
- HORT_20220038_Table_6s.xlsx
Supplementary Table 6. Information of HRM markers developed in this study
- HORT_20220038_Table_7s.pdf
Supplementary Table 7. Summary of sequence reads produced from RNA-Seq
- HORT_20220038_Table_8s.pdf
Supplementary Table 8. Information of the yuzu reference transcriptome
- HORT_20220038_Table_9s.xlsx
Supplementary Table 9. List of homologous coding sequences of single-copy genes identified from seven Citrinae species
- HORT_20220038_Table_10s.pdf
Supplementary Table 10. Statistic of mapping of trimmed reads to the reference transcriptome
- HORT_20220038_Table_11s.pdf
Supplementary Table 11. Summary of variants identified by read mapping to the reference transcriptome
- HORT_20220038_Table_12s.pdf
Supplementary Table 12. Identification of putative origins of 80 yuzu single-copy genes
- HORT_20220038_Figure_1s.pdf
Supplementary Fig. 1. Morphologies of fruits and leaves of ‘Dajungeum’ and ‘Harim12’ analyzed in this study. Scale bar indicates 1.0 cm.
- HORT_20220038_Figure_2s.pdf
Supplementary Fig. 2. Overall means of pairwise genetic distances of five hypervariable regions in chloroplast genomes of 20 Citrinae species. Species name and GenBank accession numbers of 20 species are shown in Fig. 2. Pairwise genetic distances were calculated using MEGA version X (Kumar et al., 2018). ‘Combined’ indicates a combination of all five regions.
- HORT_20220038_Figure_3s.pdf
Supplementary Fig. 3. Phylogenetic tree of Cp1 chloroplast genome sequences of yuzu and 10 papeda accessions. Detailed information of papeda accessions is shown in Supplementary Table 1.
- HORT_20220038_Figure_4s.pdf
Supplementary Fig. 4. Identification of a 1-bp InDel between two yuzu cultivars, ‘Dajungeum’ and ‘Yamane’, in the Cp1 hypervariable region of chloroplast genome sequences. A. Alignment of Cp1 sequences of two yuzu cultivars and Ichang papeda. A vertical arrow indicates the position of the 1-bp InDel. B. Chromatography of Sanger sequencing of the Cp1 region flanking the 1-bp InDel. Vertical arrow indicates the position of 1-bp InDel.
- HORT_20220038_Figure_5s.pdf
Supplementary Fig. 5. Normalized melting peak patterns of 44 high resolution melting (HRM) makers developed in this study. Detail information about HRM markers is shown in Supplementary Table 6.