Introduction
Materials and Methods
Plant materials
Conversion of single nucleotide polymorphism (SNP) markers into cleaved amplified polymorphic sequence (CAPS) markers
PCR amplification and sequencing of the PCR products
Results
Linkage relationship of closely linked molecular markers to the Ms locus
Application of the molecular markers developed for identification of cytoplasm types and Ms genotypes in onion breeding programs
Discussion
Efficient development of molecular markers using onion transcriptome sequences and rice genome information
Enhancement of efficiency of onion breeding using molecular markers developed for identification of cytoplasm types and the Ms genotypes
Introduction
Onion (Allium cepa L.) is known to originate from central Asia, but it is now cultivated all over the world through long-term breeding efforts to develop cultivars adapted to diverse environments (Brewster, 2008). Regarding the amount of world production, onion is the second most important vegetable crop, second to tomato (Griffiths et al., 2002). Onion is also known to be one of major sources of dietary flavonoids such as quercetin and anthocyanin (Fossen et al., 1996; Rhodes and Price, 1996; Slimestad et al., 2007). Although diverse landraces and open-pollinated (OP) varieties have been developed since the ancient culti-vation of onions, the proportion of F1 hybrid cultivars has been steadily increasing since the discovery of male-sterile onions by Jones and Emsweller (1936).
Male-sterility is indispensable for the production of F1 hybrid seeds in onion because self-incompatibility is not present, and mechanical emasculation is not economically feasible due to the flower structure which contains 200-600 flowers opening successively in one umbel (Brewster, 2008). Male-sterility is an inherited deficiency in producing viable pollen grains, and has been reported in more than 150 plant species (Laser and Lersten, 1972). It is classified into genic male-sterility (GMS) and cytoplasmic male-sterility (CMS) depending on the positions of the causal genes (Hanson, 1991); however, only CMS has been observed in onions. CMS has been exploited for F1 hybrid seed production in many crop species since Jones and Clarke (1943) first devised the scheme of F1 hybrid seed production utilizing CMS in onions.
CMS is known to be caused by aberrant mitochondrial genes, and several CMS-inducing genes have been isolated in crop species such as rice, maize, and radish (Budar et al., 2003; Hanson and Bentolila, 2004; Knoop, 2004; Kubo and Newton, 2008). Although almost no sequence homology has been found among CMS-inducing genes, they are mostly chimeric, consisting of partial sequences of known mito-chondrial genes along with uncharacterized sequences (Hanson and Bentolila, 2004). Such chimeric genes are assumed to be created by dynamic rearrangements of the mitochondrial genomes in plant species (Small et al., 1989; Kmiec et al., 2006), but the exact mechanism of the action by which the CMS-inducing genes lead to phenotypes of male-sterility is still unknown (Hu et al., 2014). Meanwhile, male-fertility of CMS can be restored by nuclear restorer-of-fertility (Rf) genes. Since the first cloning of the Rf2 gene in maize (Cui et al., 1996), several Rf genes have been cloned in petunia (Bentolila et al., 2002), rice (Komori et al., 2004), radish (Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003), and sorghum (Klein et al., 2005). Interestingly, most of the cloned Rf genes encode pentatricopeptide repeat (PPR) proteins. Although it is known that the PPR-coding Rf genes are targeted to mitochondria and contain RNA- binding activity, the exact mechanism by which Rf gene products suppress mitochondrial CMS-inducing genes remains unresolved (Hu et al., 2014).
Two different types of CMS have been reported in onions. The first was discovered from the cultivar ‘Italian Red’ in 1925, which is called CMS-S (Jones and Emsweller, 1936), and has been the most widely used in onion F1 hybrid breeding. Male-fertility of CMS-S is restored by a single nuclear Rf gene, the Ms locus (Jones and Clarke, 1943). Later on, another type of CMS called CMS-T was found by Berninger (1965), but the inheritance of male-fertility restoration was reported to be controlled by three inde-pendent nuclear Rf genes (Schweisguth, 1973). However, Kim (2014) proposed that fertility restoration of male-sterility conferred by both CMS-S and CMS-T cytoplasms might be controlled by the same nuclear restorer-of-fertility gene, or by two tightly linked genes. In addition, Kim (2014) hypothesized that the complex inheritance of fertility restoration of the CMS-T system proposed by Schweisguth (1973) might be caused by severe segregation distortion of the chromosomal region flanking the Ms locus.
For efficient onion F1 hybrid breeding, marker-assisted selection of the desired cytoplasm types and Ms genotypes is pivotal. Since onion is a biennial crop, it generally takes at least four years to identify the cytoplasm types and Ms genotypes through conventional progeny tests. Several molecular markers have been developed for the identification of cytoplasm type based on polymorphic sequences of the chloroplast and mitochondrial genomes (Havey, 1995; Sato, 1998; Engelke et al., 2003; Kim et al., 2009a). As for the nuclear Ms locus, although a couple of molecular makers have been developed (Gökçe et al., 2002; Bang et al., 2013, Park et al., 2013), they have experienced limitations in predicting the Ms genotypes of diverse breeding lines. However, Havey (2013) and Kim (2014) recently reported new molecular markers which were in linkage disequilibrium with the Ms locus, but the relative positions of these markers have not yet been tested. Therefore, in the present study, the relative positions of these closely linked markers to the Ms locus were analyzed, and the usefulness of the molecular markers was demonstrated for the identification of cytoplasm types and Ms genotypes in onion breeding.
Materials and Methods
Plant materials
Twenty one recombinants containing crossing overs between molecular markers closely linked to the Ms locus were used for analysis of the relative positions of the recently reported molecular markers in linkage disequilibrium with the Ms locus (Havey, 2013; Kim, 2014). The recombinants were selected from a segregating population consisting of 4,273 plants, which was produced in the previous study (Park et al., 2013). In addition, twenty-four breeding lines bred in five different onion breeding institutes in Korea were utilized to analyze the level of linkage disequilibrium between the molecular markers and the Ms locus. The cytoplasm types and male-fertility phenotypes of the breeding lines were confirmed in the previous study (Park et al., 2013).
For marker-assisted selection of homogeneous maintainer lines, 100 breeding lines which had been bred for several years in a seed company (ONBREETECH Corp., Haenam, Korea) were employed. An average of 16 seedlings of each breeding line was analyzed. The male-fertile contaminants found in the male-sterile maternal lines grown at the F1 hybrid seed production facility were analyzed to diagnose the main routes of contamination. Flower tissues of the 21 male-fertile contaminants were collected from four male-sterile parental lines, and the DNA isolated from the flower peduncles was analyzed. To test the genetic purity of the maintainer lines, two populations (‘B421’ and ‘B423’) consisting of 98 and 68 plants, respectively, were analyzed. For genetic purity tests of OP varieties, 67 and 59 onion bulbs were randomly selected from two OP varieties (‘AR13’ and ‘BS13’), and the DNA extracted from the top of each bulb was analyzed.
Total genomic DNA of the samples was extracted from the leaf, tops of the bulb, or flower peduncle tissues using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). For identification of the cytoplasm types, a previously developed molecular marker was employed (Kim et al., 2009a). This marker was developed on the basis of polymorphic mitochondrial genome organizations. The Ms genotypes of each sample were identified using the molecular marker developed by Kim (2014). This marker was shown to be in linkage disequilibrium with the Ms locus.
Conversion of single nucleotide polymorphism (SNP) markers into cleaved amplified polymorphic sequence (CAPS) markers
Havey (2013) reported three SNP markers (isotig34671_ 610, isotig30856_1351, and isotig29186_1830) which were in linkage disequilibrium with the Ms locus. To convert these markers into CAPS markers, transcriptome sequences consisting of 288,786 contigs were utilized to isolate relatively long contig sequences (unpublished). The 121-bp sequences flanking the three SNP markers presented by Havey (2013) were used as queries. Three contigs, of which the sizes were 1,358 bp, 1,722 bp, and 1,503 bp, were retrieved from the local BLAST search using BioEdit software (Hall, 1999). These contig sequences were then used to identify rice homologous genes using the Rice Genome Annotation Project database (Kawahara et al., 2013). The full-length cDNA and genomic DNA sequences were downloaded from the database. After identifying the exon and intron boundaries, the rice cDNA sequences were aligned with the onion contig sequences to identify putative exon-intron junctions in the onion contig sequences. A pair of primers was designed for the exon sequences flanking one intron. After PCR amplifi-cation, the PCR products were directly sequenced and the polymorphic recognition sites for restriction enzymes were queried to develop CAPS markers. The primer sequences and restriction enzymes used in CAPS marker genotyping are presented in Table.
PCR amplification and sequencing of the PCR products
PCR amplification of the CAPS markers developed in this study was performed in a 10 μL reaction mixture containing 0.05 μg template, 1 μL 10x PCR buffer, 0.2 μL forward primer (10 μM), 0.2 μL reverse primer (10 μM), 0.2 μL dNTPs (10 mM each), and 0.25 μL polymerase mix (Advantage 2 Polymerase Mix, Clontech, Palo Alto, CA). PCR amplification consisted of an initial denaturation step at 95°C for 4 min, 10 cycles at 95°C for 30 s, 65°C (0.8°C decrements in each cycle) for 30 s, and 72°C for 1 min, 35 cycles at 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min, and a final 10 min extension at 72°C. The PCR products were digested with their respective restriction enzymes for 3h at 37°C. The digested products were then visualized on 1.5% agarose gels after ethidium bromide staining. For sequencing of the PCR products, they were purified using a QIAquick PCR Purification kit (QIAGEN, Valencia, CA). Sequencing reactions were carried out using Big Dye (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocol, and the sequences were obtained using an ABI PRISM 3730XL Analyzer (Applied Biosystems).
Results
Linkage relationship of closely linked molecular markers to the Ms locus
Havey (2013) recently reported three SNP markers (isotig 34671_610, isotig30856_1351, and isotig29186_1830) which were in linkage disequilibrium with the Ms locus; however, the genetic distances between these SNP markers and the Ms locus, as well as the relative positions of these SNP markers in the linkage map, were not presented. To identify the linkage relationships of the three SNP markers with the Ms locus and other closely linked markers, a large segregating population, which was used in a previous study (Park et al., 2013), was analyzed.
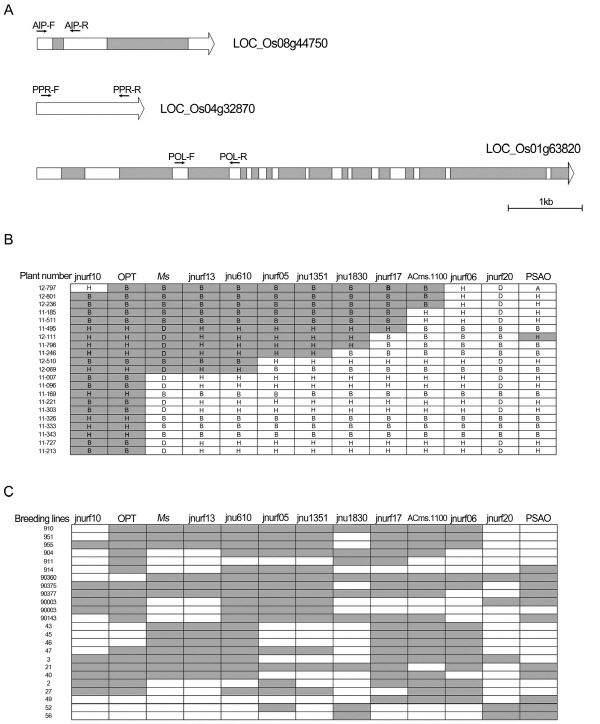
Before analysis, the three SNP markers detected by the KASPar assay (Havey 2013) were converted into readily usable CAPS markers. To develop CAPS markers based on more polymorphic intronic sequences, putative exon and intron boundaries were identified by aligning the onion contigs with rice homologs containing intron sequences. In the case of the marker isotig30856_1351, no introns were identified in the rice homolog. A number of SNPs and INDELs were identified by sequencing the PCR products, which were amplified using primer pairs designed on the adjacent exon sequences flanking an identified intron (Fig. 1A). The intron positions of the onion sequences identified by sequencing the PCR products were identical to the positions predicted by alignment with the rice homologs. Three CAPS markers, designated jnurf610, jnurf1351, and jnurf1830, were developed from the genes containing the SNP markers, isotig34671_610, isotig30856_1351, and isotig29186_1830, respectively (Table 1).
Twenty one recombinants, selected from a segregating population consisting of 4,273 plants, were analyzed using the three CAPS markers. The results showed the jnurf610 markers to perfectly co-segregate with the Ms locus together with the jnurf13 marker, while the jnurf1351 and jnurf1830 markers were positioned between the jnurf05 and jnurf17 markers (Fig. 1B). However, analysis of diverse breeding lines showed the three CAPS markers to have a discrepancy between the male-fertility phenotypes and the marker genotypes in some of the breeding lines (Fig. 1C); however, the jnurf13 marker was found to be in perfect linkage disequilibrium with the Ms locus, as shown in the previous study (Kim, 2014).
Application of the molecular markers developed for identification of cytoplasm types and Ms genotypes in onion breeding programs
Since the jnurf13 marker was found to be in complete linkage disequilibrium with the Ms locus, it would be very versatile for use in onion F1 hybrid breeding in combination with the molecular marker developed for the distinction of cytoplasm types (Kim et al., 2009a). These two molecular markers were applied to accomplish four main purposes in onion breeding programs. Among the four purposes, the selection of maintainer lines in a short period would be the most important usage of these markers.
To demonstrate the efficiency of the marker-assisted selection of maintainer lines, the two markers were used in the analysis of 100 breeding lines which had been bred for several years in a single private seed company for the purpose of developing diverse maintainer lines. An average of 16 seedling samples from each breeding line was used for analysis. The results showed most of the breeding lines to be homogeneous maintainer lines, but seven of the lines were heterogeneous, containing more than two different Ms genotypes (Table 2). For these seven segregating lines, further selection of the plants containing the homozygous recessive Ms genotype would enable the production of pure maintainer lines in the future. On the other hand, the four breeding lines harboring the homozygous dominant Ms genotype could be discarded or used as restorer lines.
In the second application, the two markers were used to diagnose the sources of male-fertile contaminants which were frequently found in the male-sterile maternal lines during F1 hybrid seed production, as shown above. These contaminants were one of the main problems causing the low genetic purity of F1 hybrid cultivars. A total of 21 male-fertile contaminants identified from four male-sterile maternal lines were analyzed with the two molecular markers. The result showed that most male-fertile contaminants were found to contain sterile cytoplasm, but their Ms genotypes were heterozygous (Table 3), suggesting that the unwanted pollen grains harboring the dominant Ms gene might have become pollinated when the male-sterile maternal lines had been produced by crossing with the maintainer lines. Meanwhile, two male-fertile off-types were shown to contain normal cytoplasm with homozygous recessive Ms genotypes. It was assumed that the seeds of these two off-types might have been introduced from maintainer lines.
As the third application, the two molecular markers were used to evaluate the genetic purity of the maintainer lines. Low genetic purity of the maintainer lines may be the major cause for the appearance of male-fertile contaminants in male-sterile maternal lines during F1 hybrid seed production. Two maintainer lines which showed a high frequency of male-fertile off-type during F1 hybrid seed production were analyzed. DNA was extracted from the leaf tissues grown from the bulbs. The bulbs were planted for seed propagation of both the maintainer lines and male-sterile parental lines. The results showed more than 10% of the individuals to contain the heterozygous Ms genotype in the ‘B421’ main-tainer line, while 15 plants were shown to have CMS-T cytoplasm (Table 4). The male-fertility phenotypes of 10 plants containing both CMS-T cytoplasm and homozygous recessive Ms genotypes were confirmed to be male-sterile after flower opening. In the case of the ‘B423’ maintainer line, all plants contained normal cytoplasm, but nine plants were found to have the dominant Ms allele (Table 4). There-fore, the presence of the dominant Ms allele in these maintainer lines must have been responsible for the frequent appearance of male-fertile off-types during F1 hybrid seed production.
Finally, the two molecular markers were applied to increase the genetic purity and seed yield potentials of OP varieties. A private seed company had been facing problems in the production of seeds of OP varieties. The number of male-sterile plants had increased every year, thus decreasing the amount of pollen grains, leading to reduced seed yields of OP varieties. The bulbs of two OP varieties, which were selected for seed propagation, were analyzed using the two molecular markers. The result showed that the proportion of bulbs containing heterozygous Ms genotypes were similar to that of bulbs possessing the homozygous dominant Ms genotype in both OP varieties (Table 5). Since the male- fertility phenotypes of the bulbs containing the heterozygous Ms genotype were male-fertile, they could not be eradicated just with visual examination. Therefore, removal of bulbs harboring the heterozygous Ms genotype using the jnurf13 marker will resolve this problem in the seed production of OP varieties.
Discussion
Efficient development of molecular markers using onion transcriptome sequences and rice genome information
Despite recent breakthroughs in genomic studies led by next-generation sequencing technologies (Hamilton and Buell, 2012), whole genome sequencing of onion seems to be intractable at least for now due to its huge genome size of 15,290 Mb per 1C nucleus (Arumuganathan and Earle, 1991), which is more than 107 times bigger than that of Arabidopsis. However, transcriptome analysis by RNA-Seq is possible in any organism, regardless of the genome size (Mutz et al., 2013; Wolf, 2013). Therefore, transcriptome sequencing in onion can provide valuable information for the development of molecular markers and construction of a high-density linkage map (Duangjit et al., 2013); however, low frequency of SNPs in the transcribed sequences and the presence of introns would be limiting factors to the development of reliable molecular markers. In many cases, synteny between the genomes of crop and model species has been used to identify orthologous genes and putative exon-intron boundaries. For example, a high-resolution linkage map of radish was constructed using the synteny between radish and Arabidopsis (Cho et al., 2012). In the case of onion, rice is the most closely related model species, but the syntenic relationship between the two genomes was reported to be very low (Martin et al., 2005; Jakše et al., 2006). This might be caused by frequent rearrangements via abundant transposable elements present in onion (Jakše et al., 2008; Vitte et al., 2013). However, the position of introns might be relatively conserved between rice and onion genes. Indeed, the positions of the introns in the two genes analyzed in this study were conserved between the two species, though the number of introns was not strictly conserved. Therefore, the streamlined framework of the rice genomics database would be a useful tool for the development of onion molecular markers.
Enhancement of efficiency of onion breeding using molecular markers developed for identification of cytoplasm types and the Ms genotypes
Since the first discovery of a male-sterile onion by Jones and Emsweller (1936), the proportion of F1 hybrid cultivars in onion has increased worldwide, as for other major crops. Except for a few OP cultivars, F1 hybrids have been pre-dominantly cultivated in Korea as well. Male-sterility is the sole tool for the economical production of F1 hybrid seeds in onions, as both self-incompatibility and mechanical emasculation are not applicable in onions. However, the development of male-sterile maternal and its isogenic male- fertile maintainer lines are required for the production of F1 hybrid seeds using male-sterility. To be used as a maintainer line, the cytoplasm type must be normal, and the genotype of the restorer-of-fertility gene should be recessive homozygous. Through convention progeny tests, it usually takes at least four years to identify the cytoplasm types and the Ms genotypes in onion due to its biennial generation time. In addition, the fact that progeny tests are laborious and demand large space is a limiting factor for the development of diverse elite maintainer lines.
However, such a limiting factor can be overcome by the use of molecular markers distinguishing the cytoplasm types and Ms genotypes, thus enabling dramatic enhancement of the efficiency of F1 hybrid breeding. In this study, it was demonstrated that these molecular markers could be effectively used in onion breeding programs. Homogeneous maintainer lines were successfully selected from candidate breeding lines in a significantly shorter period compared with progeny tests. In the second application, the plausible sources of male-fertile off-types found in male-sterile maternal lines during F1 hybrid seed production were successfully diagnosed using the two molecular markers. Consequently, contamination with pollen grains containing the dominant Ms allele was thought to be the major route responsible for the appearance of the male-fertile off-types in this study. However, inadvertent seed contamination can also be a major source depending on the different situations in each seed company. Therefore, these molecular markers can be utilized in assessing the possible routes of appearance of male-fertile off-types and preventing their appearance during F1 hybrid seed production. In addition, high genetic purity of the maintainer lines was fundamentally important to prevent the appearance of male-fertile off-types. Therefore, it is recommended that breeders or managers in charge of seed propagation periodically test the genetic purity of the maintainer lines using these two molecular markers. This practice will assure production of high-quality F1 hybrid seeds.
The Ms genotypes of all samples analyzed in this study were successfully predicted using the jnurf13 marker, which was in linkage disequilibrium with the Ms locus (Kim, 2014). As shown in the previous study (Kim, 2014), the jnurf13 marker could be used in both CMS-S and CMS-T systems in this study. This supports the hypothesis that male-sterility induced by CMS-S and CMS-T cytoplasms can be reversed by the same or tightly linked restorer-of-fertility genes (Kim, 2014). Therefore, as far as this hypothesis is true, it would be unnecessary to distinguish between CMS-S and CMS-T cytoplasms in onion breeding programs. This fact would be very helpful for onion breeders in Korea, since the frequency of CMS-T cytoplasm was reported to be more than three times higher than that of CMS-S cytoplasm in F1 hybrid cultivars bred in Korea (Kim et al., 2009b). For some seed companies, almost all male-sterile lines used in F1 hybrid breeding contained the CMS-T cytoplasm (data not shown). Although diverse F1 hybrid cultivars bred in other countries could not be tested in this study, the male- fertility phenotypes and the Ms genotypes predicted by the jnurf13 marker were perfectly matched in some F1 hybrid cultivars bred in Europe (data not shown). However, more extensive tests of the applicability of the jnurf13 marker in diverse onion F1 hybrid cultivars and breeding lines bred all over the world will be necessary in the future.