Introduction
Materials and Methods
Oryzalin Treatment of the Experimental Materials
Flow Cytometry Analysis
Morphological Analysis
Chromosome Analysis
Results and Discussions
Flow Cytometry Analysis of the Ploidy Level
Morphological Analysis
Chromosome Analysis
Introduction
Cucumis melo ‘Chammel’ is a new cultivar from a cross between oriental melon and melon. It has an orange pulp, similar to a melon, and a shape similar to an oriental melon. It has high sugar, citric acid, and beta-carotene contents, and a crisp taste. (Bae et al., 2019). Oriental melons are susceptible to powdery mildew, which frequently infects Cucurbitaceae plants (Lee et al., 2010) while, Cucumis melo ‘Chammel’ displays robust resistance to this disease (Bae et al., 2019).
Plant size, disease resistance, environmental stress tolerance, and seedlessness of fruits are all desirable traits in horticultural crops that are introduced by breeding and polyploidization (Ranney, 2006). Consumers prefer seedless fruits because of their higher sugar content (Marr and Gast, 1991), superior consistency, and higher yield (Henderson, 1977). As a result, one of the primary objectives of modern breeding is seedlessness. Seedless triploid watermelons are created by first generating tetraploids with a chromosome doubling reagent and then crossing the tetraploids with diploids (Suying et al., 1993; Jaskani et al., 2004).The most common chromosome doubling reagent is colchicine (C22H25NO6) (Dermen, 1940). However, colchicine is carcinogenic and can cause infertility and chimerism (Wan et al., 1989). Therefore, the use of colchicine has been limited. Another chromosome doubling reagent, oryzalin (3, 5-dinitroN4, N4-dipropylsulfanilamide) has been successfully used to induce polyploidization in crops (Chauvin et al., 2005). Oryzalin is a synthetic herbicide used for weed control and inhibits microtubule assembly during cell division. Oryzalin has a higher affinity for plant tubulins (Doležel et al., 1994), and usually achieves the best results of polyploidization when young, actively growing meristems are treated (Ascough et al., 2008).
During polyploidization experiments, the change in nuclear DNA content needs to be monitored using various methods. For example, flow cytometry offers a quick, precise and simple way to determine nuclear DNA content (C-value) (Galbraith et al., 2009). In addition, fluorescence in situ hybridization (FISH) and karyotyping are commonly used techniques for cytogenetic studies (Anamthawat-Jónsson, 2004; Hwang et al., 2020; Islam et al., 2020). FISH analysis may provide basic details about each species’ ploidy degree and chromosomal characteristics. Furthermore, the 5S, 18S, 25S, and 45S rDNA loci have been widely used to investigate the genetic relationships among plant species. (Pellerin et al., 2019; Sakhanokho et al., 2020). This study aimed to understand the effects of oryzalin on the polyploidization of Cucumis melo ‘Chammel’ with the goal of creating new seedless oriental melon cultivars.
Materials and Methods
Oryzalin Treatment of the Experimental Materials
Fifty-eight diploid seedlings of oriental melon (Cucumis melo) ‘Chammel’ with high-sugar content were used in this experiment. They were grown in a greenhouse (Spring Seed Co., Ltd, Korea) with day temperatures maintained between 25 to 30°C and night temperatures between 20 to 25°C. One week-old seedlings were treated with 50 mg․L-1 of oryzalin;10 µL was dropped at the shoot apex of young leaves immediately after cotyledons opened. The cotyledons were fixed with clips for 7 days to increase the absorption of oryzalin.
Flow Cytometry Analysis
The young leaves of treated plants were collected for flow cytometric analysis (Partec PA, Ploidy Analyzer, and Germany). The leaves were cut into (0.5 cm2) pieces, put in a 5 × 12 mm Petri dish, and chopped with a blade in 500 µL of nuclei isolation buffer. Then, solution was filtered into a tube to remove the debris. Next, 2 ml of DAPI staining buffer (Partech, GmbH, Munster, Germany) was poured into the tube. The nuclei suspension was then introduced into a flow cytometry analyzer. After checking the ploidy level, the tetraploid plants were selected for self-crossing and seed collection for further cultivation.
Morphological Analysis
Seed Observation
The seed morphology of the tetraploid plants was compared to that of the diploid plants. The length, width, thickness, and weight of the seeds were measured.
Leaf Observation
The morphology of the cotyledon and the first true leaves diploid and tetraploid plants was observed. The cotyledon length and width; and the length, width, thickness of first leaves, and the length of petiole were measured.
Stomata Observation
The morphological features of the stomata of the diploids and tetraploids were compared. The epidermis was separated using forceps, placed on a glass slide, and observed under a microscope (BX61, Olympus, Japan) at 200X magnification.
Chromosome Analysis
Slide Preparation
Root tips of tetraploid and diploid plants were collected early in the morning, treated with 2mM 8-hydroxyquinoline at 20°C for 4 hours, and fixed overnight in a solution (ethanol: acetic acid=3:1). The root tips were then put in a refrigerator with 70% ethanol. They were washed with distilled water before being treated an enzyme solution of 0.3% pectolyase, 0.3% cellulase, and 0.3% cytohelicase in 150 mM citrate buffer for 40 minutes at 37°C. The root-tips were then squashed in 17 µL of 70% acetic acid on a glass slide and air-dried. Under a fluorescence microscope, the chromosomes were stained with 6-diamino-2-phynylindole (DAPI) in VECTASHIELD solution and counted (BX 61, Olympus, Japan).
FISH Treatment
FISH analysis was conducted according to Lim et al. (2001) with a minor modification. First, DAPI was removed from the slides using 2X SSC buffer for 10 min. Then, the slides were dehydrated by treating for 9 min each with 70%, 90%, and 100% ethanol. Briefly, slides were pre-treated with 100 µg·mL-1 RNase A at 37°C for 1 hour. After rinsing them with 2× SSC and fixing them with 4% paraformaldehyde for 10 min, we again rinsed the slides with 2× SSC and dehydrated them in an ethanol series (70%, 90%, and 100%), followed by air-drying.
The hybridization mixture contained formamide, 50% dextran sulfate, and 20 × SSC, 10% sodium dodecyl sulfate, herring sperm DNA, and rDNA probes. The mixture was placed in a water bath for DNA denaturation at 82°C for 2 min and then put on ice for 15 min for fixation. Forty microliters of the mixture were added to each slide, and a coverslip was placed on the slide, while ensuring there were no bubbles. The slides were put in a water bath for hybridization at 82°C for 2 min and then placed in a container with wet tissue paper and incubated in a humid chamber at 37°C for 18-20 hours. The slides were washed with 2× SSC buffer for 5 min followed by 0.1×SSC buffer for 30 min at 42°C with shaking and 2×SSC buffer for 5 min. Then, the slides were submerged into 1× detection buffer for 5 min. Streptavidin CY3 (Invitrogen, Carlsbad, CA, USA) and anti-digoxigenin fluorescein (Roche, Basel, Switzerland) were used to detect the labeled chromosomes. The slides were placed with coverslips in a humid chamber and incubated at 37°C for 1 hour. Three jars were incubated containing a 1× detection buffer in the dark in a water bath at 37°C for 5 min, followed by dehydration in an ethanol series (70%, 90%, and 100%). Then, the slides were air-dried in the dark and counterstained with DAPI and Vectashield at a 2:100 ratio (Vector Laboratories, Burlingame, CA, USA).The prepared slides were examined with a model Nikon BX 61 fluorescence microscope (Tokyo, Japan). The probe signals were analyzed using ultraviolet excitation filters. Cytovision and imaging software were used to acquire the chromosomes’ images with the 5S rDNA and 18S rDNA signals.
Statistical Analysis
Each of the characteristics was compared between the diploids and tetraploids using the SAS package (Statistical Analysis System, version 9.4, SAS Institute Inc.). The phenotypic similarity was analyzed using a 1% significance level in a t-test for a completely randomized design.
Results and Discussions
Flow Cytometry Analysis of the Ploidy Level
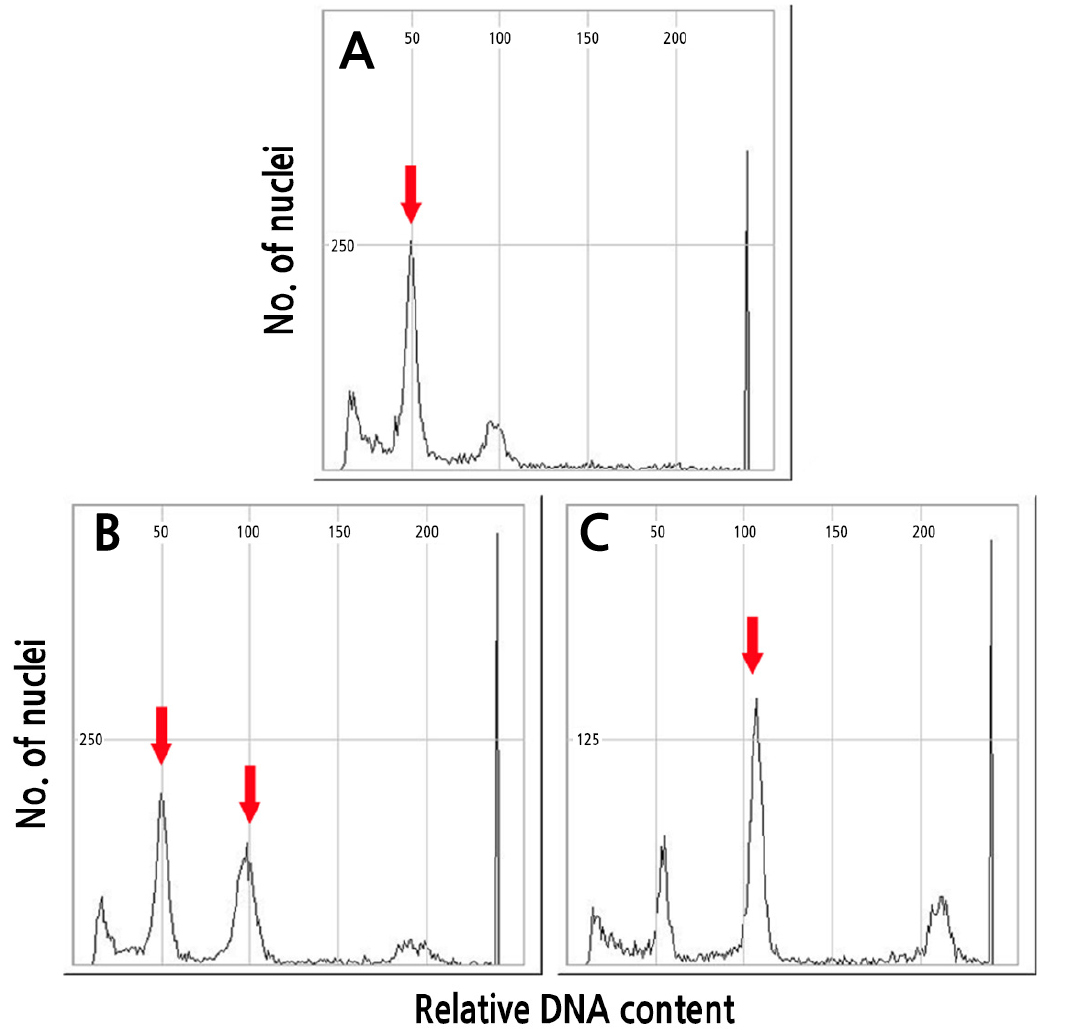
The DNA content of the 58 oryzalin-treated Cucumis melo ‘Chammel’ plants were analyzed by flow cytometry. The ploidy levels of were determined by the peak’s channel position. An untreated diploid ‘Chammel’ plant was used as the control with a peak observed at channel 50 (Fig. 1A). The peak of a tetraploid line was observed at channel 100 (Fig. 1C), and the peaks of a mixoploid line were observed middle positions at channels 50 and 100 (Fig. 1B). Among the 58 plants diploid oryzalin treated ‘Chammel’ plants, 12 were detected as tetraploids, one was mixoploid, and the remaining were diploid (Table 1). In the case of watermelon treated by oryzalin, they found diploids, mixoploids, and desired tetraploids. (Kwack and Lee, 1999; Bae et al., 2020). In a study by Raza et al. (2003) using 0.01% colchicine in watermelon, the highest number of tetraploids was found. Tetraploid plants can be used to produce seedless nursery crops by creating triploids (Contreras et al., 2007). Therefore, these tetraploid oriental melons could be used for breeding with diploids and make new seedless oriental melon cultivars.
Table 1.
The ploidy level of the 58 seedlings of Cucumis melo ‘Chammel’ treated with 50 mg․L-1 oryzalin
| Plants | Mean | Ploidy | 2C DNA content (Mbp) | pg (2C)z |
| Control | 52.30 | 2x | 442.46 | 0.45 |
| OM1 | 110.08 | 4x | 931.28 | 0.95 |
| OM2 | 57.60 | 2x | 487.30 | 0.50 |
| OM3 | 56.27 | 2x | 476.04 | 0.49 |
| OM4 | 109.59 | 4x | 927.13 | 0.95 |
| OM5 | 54.35 | 2x | 459.80 | 0.47 |
| OM6 | 55.64 | 2x | 470.71 | 0.48 |
| OM7 | 56.30 | 2x | 476.30 | 0.49 |
| OM8 | 57.25 | 2x | 484.34 | 0.50 |
| OM9 | 106.58 | 4x | 901.67 | 0.92 |
| OM10 | 107.24 | 4x | 907.25 | 0.93 |
| OM11 | 105.64 | 4x | 893.70 | 0.91 |
| OM12 | 55.47 | 2x | 469.28 | 0.48 |
| OM13 | 103.70 | 4x | 877.30 | 0.90 |
| OM14 | 100.46 | MIX | 849.89 | 0.87 |
| OM15 | 105.02 | 4x | 888.47 | 0.91 |
| OM16 | 54.64 | 2x | 462.25 | 0.47 |
| OM17 | 54.59 | 2x | 461.83 | 0.47 |
| OM18 | 55.85 | 2x | 472.49 | 0.48 |
| OM19 | 54.34 | 2x | 459.72 | 0.47 |
| OM20 | 55.50 | 2x | 469.53 | 0.48 |
| OM21 | 105.13 | 4x | 889.40 | 0.91 |
| OM22 | 55.70 | 2x | 471.22 | 0.48 |
| OM23 | 53.93 | 2x | 456.25 | 0.47 |
| OM24 | 103.70 | 4x | 877.30 | 0.90 |
| OM25 | 51.74 | 2x | 437.72 | 0.45 |
| OM26 | 52.69 | 2x | 445.76 | 0.46 |
| OM27 | 53.90 | 2x | 455.99 | 0.47 |
| OM28 | 104.00 | 4x | 879.84 | 0.90 |
| OM29 | 52.12 | 2x | 440.94 | 0.45 |
| OM30 | 51.34 | 2x | 434.34 | 0.44 |
| OM31 | 54.03 | 2x | 457.09 | 0.47 |
| OM32 | 52.28 | 2x | 442.29 | 0.45 |
| OM33 | 54.66 | 2x | 462.42 | 0.47 |
| OM34 | 55.05 | 2x | 465.72 | 0.48 |
| OM35 | 52.55 | 2x | 444.57 | 0.45 |
| OM36 | 54.57 | 2x | 461.66 | 0.47 |
| OM37 | 54.36 | 2x | 459.89 | 0.47 |
| OM38 | 53.28 | 2x | 450.75 | 0.46 |
| OM39 | 53.66 | 2x | 453.96 | 0.46 |
| OM40 | 53.30 | 2x | 450.92 | 0.46 |
| OM41 | 54.72 | 2x | 462.93 | 0.47 |
| OM42 | 53.12 | 2x | 449.40 | 0.46 |
| OM43 | 53.76 | 2x | 454.81 | 0.47 |
| OM44 | 53.32 | 2x | 451.09 | 0.46 |
| OM45 | 54.51 | 2x | 461.15 | 0.47 |
| OM46 | 51.79 | 2x | 438.14 | 0.45 |
| OM47 | 53.22 | 2x | 450.24 | 0.46 |
| OM48 | 54.01 | 2x | 456.92 | 0.47 |
| OM49 | 52.14 | 2x | 441.10 | 0.45 |
| OM50 | 102.84 | 4x | 870.03 | 0.89 |
| OM51 | 54.16 | 2x | 458.19 | 0.47 |
| OM52 | 53.20 | 2x | 450.07 | 0.46 |
| OM53 | 55.47 | 2x | 469.28 | 0.48 |
| OM54 | 52.09 | 2x | 440.68 | 0.45 |
| OM55 | 51.39 | 2x | 434.76 | 0.44 |
| OM56 | 53.69 | 2x | 454.22 | 0.46 |
| OM57 | 53.82 | 2x | 455.32 | 0.47 |
| OM58 | 101.01 | 4x | 854.54 | 0.87 |
z1 Mbp = 1/978 pg according to Doležel et al. (2007).
Morphological Analysis
Observation of Seeds
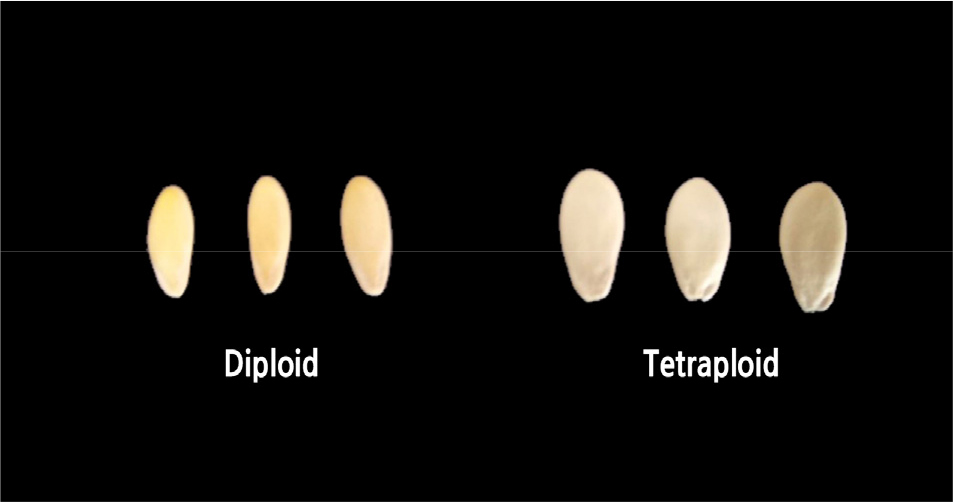
The tetraploid ‘Chammel’ seeds had average width of 4.65 mm and average weight of 31.82 mg, while the diploids had average width of 3.61 mm and average weight of 21.18 mg. (Table 2 and Fig. 2). However, the tetraploid and diploid seeds had similar lengths and thicknesses (Table 2). Previous studies found that polyploid seeds of Hylocerous (Cohen et al., 2013), Japanese dandelion (Hoya et al., 2007), and Dactylis glomerata (Maceira et al., 1993) were larger than the corresponding diploids.
Table 2.
The comparison of seed morphology between Cucumis melo ‘Chammel’ diploids and tetraploids
| Ploidy level | Length (mm) | Width (mm) | Thickness (mm) | Weight (mg) |
| Diploid | 9.25 ± 0.13 | 3.61 ± 0.06 | 1.61 ± 0.02 | 21.18 ± 0.25 |
| Tetraploid | 9.39 ± 0.13 | 4.65 ± 0.05 | 1.62 ± 0.04 | 31.82 ± 0.18 |
| Significance | NS | ** | NS | ** |
Observation of Leaves
Tetraploids had average cotyledon width of 2.05 cm and a first true leaf thickness of 0.64 mm, while diploids had average cotyledon width of 1.60 cm and a first true leaf thickness of 0.31 mm. (Table 3 and Fig. 3). A similar pattern was found in Lycopersicon esculentum, in which leaves of the tetraploids were broader than those of the diploids (Ishfaq et al., 2012). However, the petiole length of the tetraploid (3.42) cm was shorter than that of the diploid (4.87 cm), and significant difference were found in the length and width of the first true leaf between the diploids and tetraploids (Table 3). The difference at the leaf margin was noticeable. The leaf margin of the tetraploids was crenate, and the entire margin was smooth in the diploids (Fig. 3). The leaves of polyploid plants often show several distinct characteristics, such as thicker leaf lamina and shape changes (Eng and Ho, 2019).
Table 3.
The comparison of leaf morphology between Cucumis melo ‘Chammel’ diploids and tetraploids
| Ploidy level | Cotyledon | First true leaf | ||||
| Length (cm) | Width (cm) | Length (cm) | Width (cm) | Thickness (mm) | Petiole length (cm) | |
| Diploid | 3.17 ± 0.03 | 1.60 ± 0.07 | 5.65 ± 0.04 | 6.35 ± 0.08 | 0.31 ± 0.01 | 4.87 ± 0.08 |
| Tetraploid | 3.17 ± 0.03 | 2.05 ± 0.04 | 5.03 ± 0.09 | 6.78 ± 0.05 | 0.64 ± 0.02 | 3.42 ± 0.06 |
| Significance | NS | ** | ** | ** | ** | ** |
Observation of Stomata
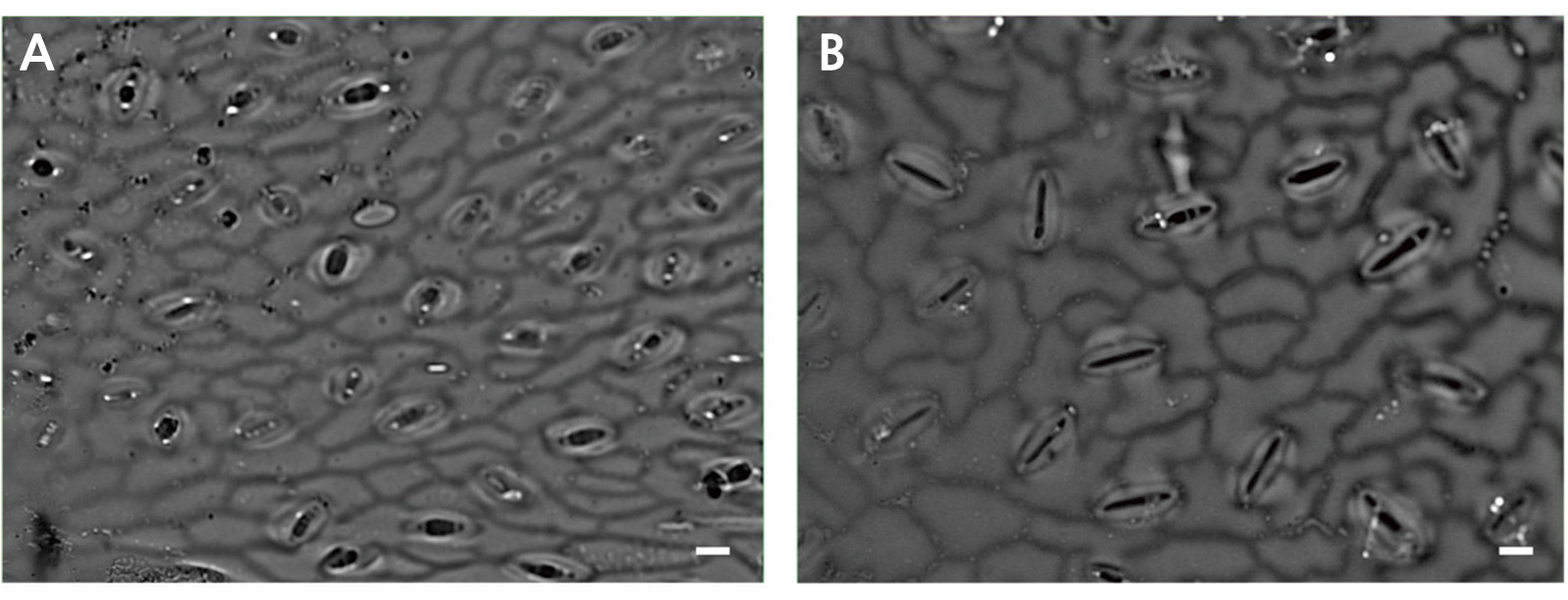
The guard cell’ length and width and the stomata’ density of the diploids and tetraploids were measured (Table 4 and Fig. 4). The average length and width of the guard cells were 48.67 µm and 6.06 µm, respectively, for the tetraploids, and 32.04 µm and 3.67 µm, respectively, for the diploids. Polyploidy can be easily measured by visualizing the guard cells since tetraploid stomata become larger than diploid stomata (Yang et al., 2006). In this study, the tetraploid leaves had a stomatal density of 14 cell/mm2, while diploid leaves had nearly twice the density at 26.67 cell/mm2. This finding is similar to the results from watermelon (Bae et al., 2020) and coffee (Mishra, 1997). The polyploid plants may increase stress tolerance by reducing transpiration, such as the drought-tolerant autopolyploid in Arabidopsis (Del Pozo and Ramirez-Parra, 2015) and drought-tolerant allopolyploids in wheat (Xiong et al., 2006).
Table 4.
The comparison of stomata distribution and size between Cucumis melo ‘Chammel’ diploids and tetraploids
| Ploidy level | Stomata | Guard cell | |
| Density (cell/mm2) | Length (µm) | Width (µm) | |
| Diploid | 26.67 ± 0.88 | 32.04 ± 3.44 | 3.67 ± 0.26 |
| Tetraploid | 14.00 ± 0.58 | 48.67 ± 0.94 | 6.06 ± 0.25 |
| Significance | ** | ** | ** |
Chromosome Analysis
Chromosome Doubling
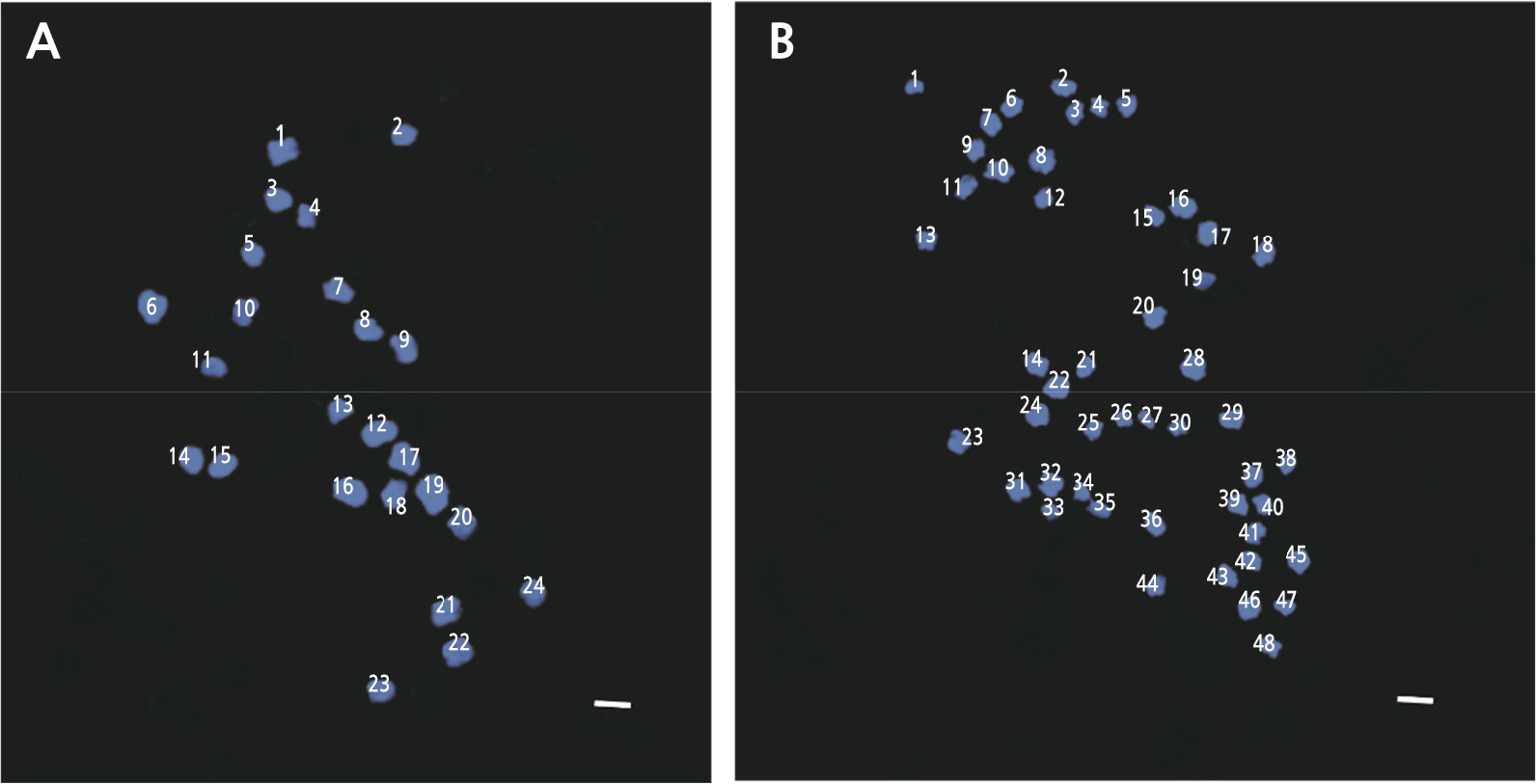
Determining chromosome numbers is a fundamental tool in cytogenetic studies (Mohammad et al., 2020). Korean melon is a diploid plant with a basic chromosome set (x) of 12 (2n = 2x = 24) (Kim et al., 2016). In this study, we found that the diploid Korean melon, also the control, had 24 (2n = 2x = 24) chromosomes, and the tetraploid oriental melon ‘Chammel’ plants had 48 chromosomes, (2n = 4x = 48) (Fig. 5). This result is consistent with the study of watermelon polyploidization in which diploid (2n = 2x = 22) plants treated with oryzalin became tetraploid (2n = 4x = 44) (Bae et al., 2020), and chromosome doubling of Hibiscus such as H. syriacus ‘Oiseau blue’ and ‘Woodbrize’ in which young leaves with colchicine generated polyploid cultivars (Van Laere et al., 2006).The effective induction of chromosome doubling in diploid and triploid roses, by oryzalin has been documented (Kermani et al., 2003). In agriculture, several polyploid plants, e.g., wheat, cotton, coffee, oat, and canola, are widely cultivated, suggesting that polyploidy is very useful (Zeng et al., 2019).
FISH Analysis
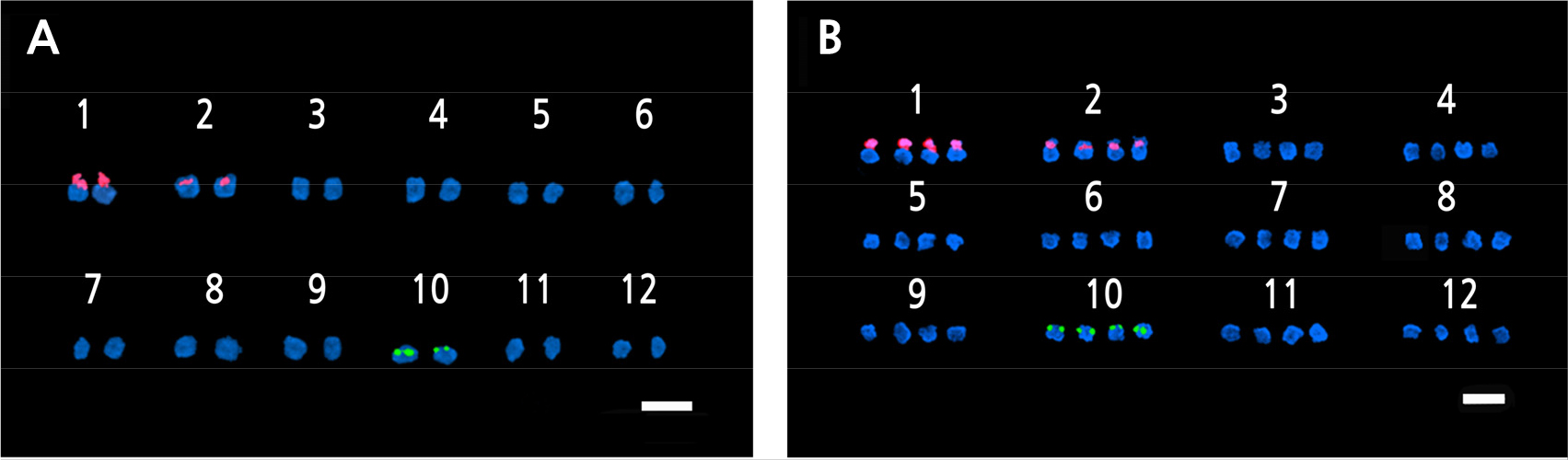
FISH is a molecular cytogenetic technique that uses fluorescently labeled probes to determine the presences of a particular DNA sequence in the nuclei (Speicher and Carter, 2005; Hwang et al., 2020). FISH analysis was used for the accurate confirmation of ploidy in a previous study (Islam et al., 2020; Waminal and Kim, 2015). The 5S rDNA and 18S rDNA loci are usually located at different sites on different chromosomes, and their transcription is carried out by rRNA polymerases. These markers are widely used in FISH techniques. Frequently used 5S rDNA loci includes the 5S rRNA repeated units plus intergeneric spacer regions (IGS) and frequently used 45S rRNA loci include the 18S, 5.8S, and 25S rDNA repeated units, internal transcribed sequences (ITS1 and ITS2), and the IGS spacer region (Volkov et al., 2017). According to the FISH results, one pair of 5S rDNA loci and two pairs of 18S rDNA loci were detected in the diploid oriental melons, whereas two pairs of 5S rDNA and four pairs of 18S rDNA loci were detected in the tetraploid melons (Fig. 6). A study of cotton plants revealed that most diploids had one pair of 5S rDNA loci, and all the allotetraploid species had two pairs (Gan et al., 2013); the same result was found in the woody species of the genus Rubus (Wang et al., 2015). The 5S rDNA loci copy number, which was found in most FISH studies, is generally used to determine the ploidy level of plants (Younis et al., 2015; Mohammad et al., 2020).
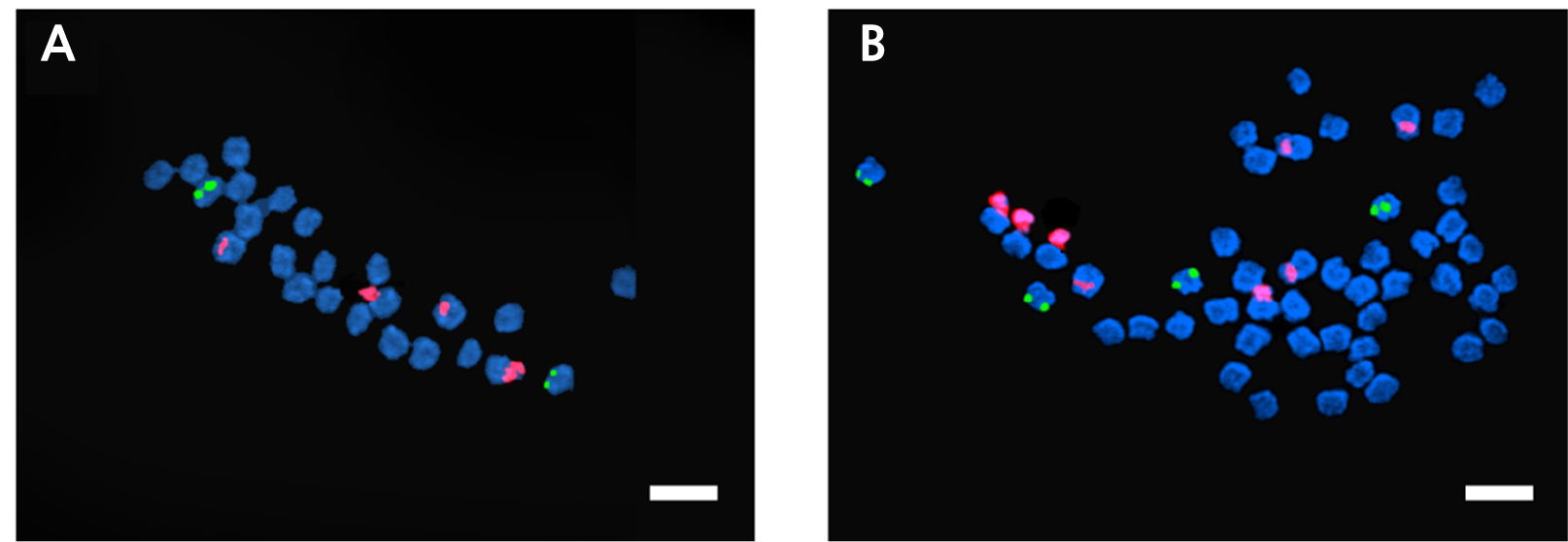
Fig. 6.
Fluorescence in situ hybridization (FISH) images of 5S and 18S rDNA-labeled metaphase chromosomes of Cucumis melo ‘Chammel’ plants. A. Diploid (2n = 2x = 24).B. Tetraploid (2n = 4x = 48). The green and red fluorescence indicate the 5S and 18S rDNA loci, respectively. Magnification: 1000×. Scale bar = 10µm.
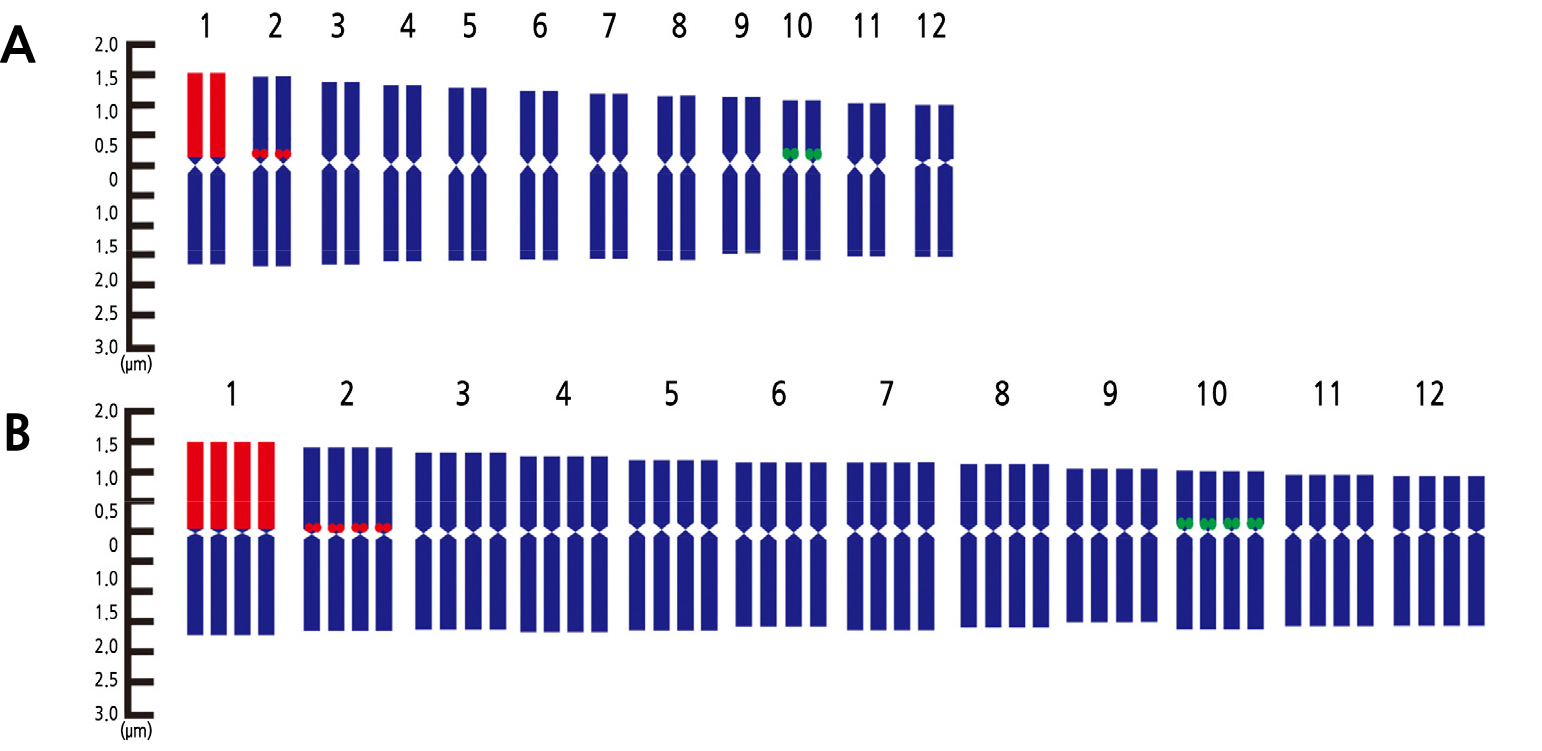
Karyotyping is a technique for detecting chromosome variation and rDNA loci distributions, as well as creating a genetic map (Chung et al., 2018). It is the most used cytological technique in empirical and theoretical studies (Liang and Chen, 2015). In this study, 18S rDNA signals were found in chromosome 1 and 2 for both the diploids and tetraploids. In the case of 5S rDNA, its signals were detected near the centromere of chromosome 10 for both the diploids and tetraploids (Figs. 7 and 8). The FISH and karyotype data confirm the polyploidization of the ‘Chammel’ plants. This polyploidization method is expected to allow for more hybridizations with diploid oriental melons, allowing for the development of seedless cultivars.