Introduction
Materials and Methods
Materials
Root collection
Slide preparation
FISH
Results
Chromosome counts and length
5S and 18S ribosomal DNA sites
Chromosome karyotyping
2C-DNA content
Discussion
Introduction
The melon Cucumis melo, which grows worldwide, has a diploid (2n) chromosome count of 24 and is an important fruit of the Cucurbitaceae family. Melons are remarkably diverse in terms of their color, size, peel, and flesh (Xu et al., 2022). The cantaloupe melon is a muskmelon variety that weighs 0.5–5 kg (Pitrat, 2016). The netted melon, which is the best-known and most widely consumed melon worldwide, has many varieties that stand out owing to its round shape, colorful peel, flesh, and texture. Hami melons have an elongated oblong shape with rounded and curved ends. These melons have a substantial amount of sugar, resulting in a rejuvenating sweet and fruity taste with hints of tropical and honey-like flavors. Mature melons have a distinct pleasant aroma, which, along with their sweetness, is a primary attribute (Chakravarty, 2019). Green or yellow green melon flesh usually contains a significant amount of vitamin C, whereas red flesh is rich in vitamin A due to its high amount of carotene (Choi et al., 2010; Liu et al., 2010; Li et al., 2024). Melons are high-quality fruits with significant nutritional value (Joo et al., 1996), and they are very popular worldwide.
In the melon industry, breeding is necessary for increased fruit quality, pest resistance, and productivity. To select the desired parents and obtain the expected offspring, cytogenetic characteristics such as the chromosomal number and length, rDNA loci, genome size, and putative DNA content should be known before breeding. Fluorescent in situ hybridization (FISH) and karyotyping are highly effective cytogenetic techniques (Anamthawat-Jónsson, 2004; Nihat et al., 2021). FISH can provide information about ploidy and chromosomal features (Sakhanokho et al., 2020). The present study aims to determine the karyotypes of three melon cultivars during the somatic metaphase using 5S and 18S ribosomal DNA. These techniques enable us to distinguish between each chromosome’s short and long arms and the unique somatic chromosomes of the cultivars. For a deeper understanding of the evolutionary process of Cucumis melo (C. melo), the spatial arrangements of these distinctive melon chromosome features were analyzed. Furthermore, flow cytometry allows rapid, accurate, and uncomplicated assessments of the nuclear plant DNA concentration (C-value). This method simplifies plant species classification in natural and agricultural contexts and also enables easy determination of plant ploidy, which is influenced by the surrounding conditions (Galbraith, 2009). This study sought to establish a foundation for cytogenetic investigations of cantaloupe, netted, and Hami melons.
Materials and Methods
Materials
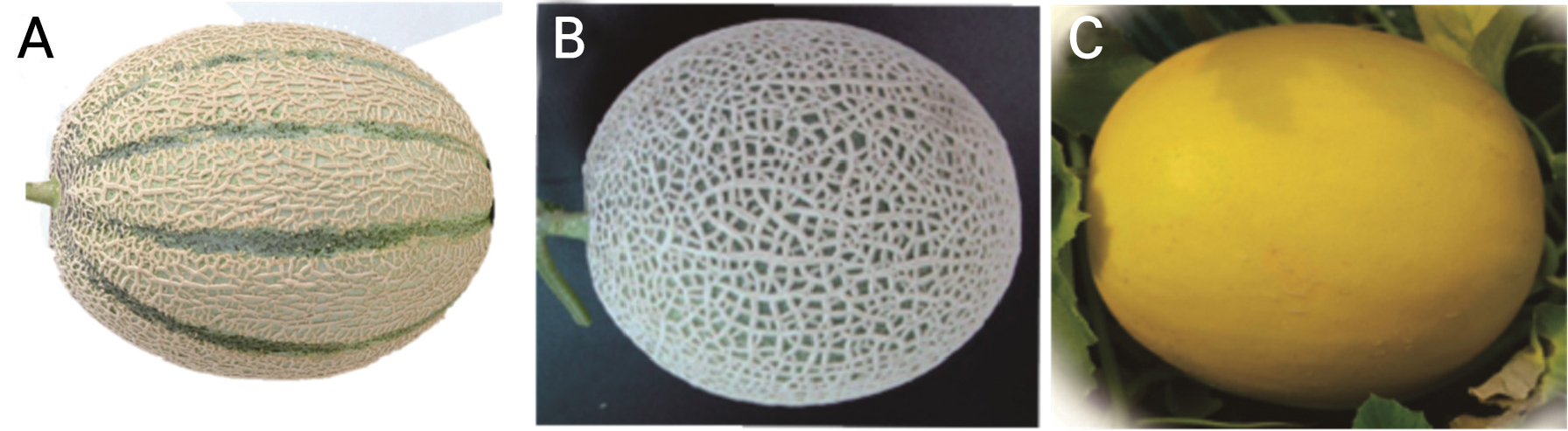
C. melo var. cantalupensis ‘JCCM-02’, C. melo var. reticulatus ‘Top Earl’s’, and C. melo var. saccharinus ‘Hami Gold’ seedlings were obtained from the Jangchun seed company in Korea 30 days after sowing (Fig. 1). The seedlings were grown and nurtured at a smart farm managed by Kyungpook National University in Daegu, South Korea, under proper fertigation and atmospheric conditions.
Root collection
Melon root samples were collected at 7 am and then placed in sun-protected tubes containing 2 mM 8-hydroxyquinoline at room temperature for four hours. Next, they were treated with a 3:1 ethanol–acetic acid solution for 24 hours. The root tips were then washed with distilled water and preserved in 70% ethanol at ‒20°C until use.
Slide preparation
To remove the fixative, the root tips were washed using distilled water. Next, 0.7–1.0-mm samples were obtained from the distal tip of the root, where the actively dividing meristem is located. Next, four to five root meristems were transferred to 1.5 mL tubes containing 60 µL of an enzyme mixture containing 1% cellulase, 1% pectolyase (Seishin Pharmaceutical Co., Li, Japan), and 1% cytohelicase in 0.01 M citrate buffer and incubated at 37°C for one hour. The digestion time can vary depending on several variables, including the root tip size and the age of the enzyme. After incubation, the tubes were placed on ice for five min. Next, the supernatants were carefully discarded using a pipette to prevent damage to the digested root meristems at the bottom. Carnoy’s solution (3:1) was then added to the tubes, followed by homogenization of the treated roots. The tubes containing the digested root meristems were briefly vortexed to obtain a cell suspension and were then placed on ice for five min. They were subsequently centrifuged at 12,000 rpm for two min, and the supernatants were discarded by pipetting. Pellets were then resuspended in 20–30 µL of an acetic acid–ethanol (9:1) solution. Slides were placed in a modified steam generator (water bath at 80°C) until the surface appeared granule-like (i.e., an ethanol meniscus formed on the slide, 10–15 S). Next, 10 µL of the cell suspension was placed on the moistened slides, which were then dried for further experiments.
FISH
Pre-treatment
The FISH protocol published by Lim et al. (2001) was used, with minor changes. Slides were treated with a solution containing 98 µg·mL-1 RNase A and 2 ml of 2X SSC buffer at 37°C for 45 min. They were then washed with 2X SSC buffer for 15 min and rinsed with 4% paraformaldehyde for 10 min. The slides then underwent a series of rinses with 70%, 90%, and 100% ethanol for nine min before being air-dried.
Hybridization
5S rDNA and 18S DNA oligoprobes were used for the FISH analysis. A solution of pre-labeled 5S rDNA and 18S rDNA was prepared by mixing with formamide, 50% dextran sulfate, and 20X SSC. Being careful to avoid bubbles, 40 µL of the solution was placed on each slide. For hybridization, the slides were incubated at 82°C for five minutes in a water bath. They were then incubated for 45 min at 37°C in a growth chamber. Again, a 42°C water bath (with shaking) was utilized for the slides with 0.1X SSC buffer, for 30 min. Finally, the slides were washed using a 70%, 90%, and 100% ethanol series.
Detection
The slides were stained (in the dark) with a solution containing (4',6-diamidino-2-phenylindole) DAPI and Vectashield (Vector Laboratories, Burlingame, CA, USA) at a ratio of two parts DAPI to 100 parts Vectashield and were subsequently examined under a Nikon BX 61 fluorescent microscope (Tokyo, Japan). The 5S and 18S rDNA signals were detected and captured using the Cytovision software.
Karyotyping
Cells exhibiting adequate metaphase chromosome dispersions were selected for karyotyping. The Cytovision software was used to determine the chromosomal arm length. Next, the chromosomes were arranged based on length of the short arm and were assigned a numerical designation (Lim et al., 2001). The classification was based on the ratio of the arms’ lengths, according to Levan (1964).
Flow cytometry
Young melon leaves were used for flow cytometry (Fig. 2), which was done on a Partec PA ploidy analyzer (Germany). The samples were cut into 0.5 × 0.5 cm fragments, placed in a 50 × 12-mm petri dish, and then finely minced using a blade. Next, 500 µL of a nucleus isolation buffer was added. The crushed leaf samples were then filtered into a tube to remove any unwanted particles, with this followed by the addition of 2 mL of DAPI staining buffer (Partech, GmbH, Munster, Germany). The nuclear suspension was then transferred to flow a cytometer. The 2C genome sizes were determined by multiplying the DNA content of a standard by the mean fluorescence value of the sample divided by the standard’s mean fluorescence value (Doležel et al., 1992). Raphanus sativus ‘Saxa’ (2C genome size: 1.10 pg) was used as the reference genome size standard.
Results
Chromosome counts and length
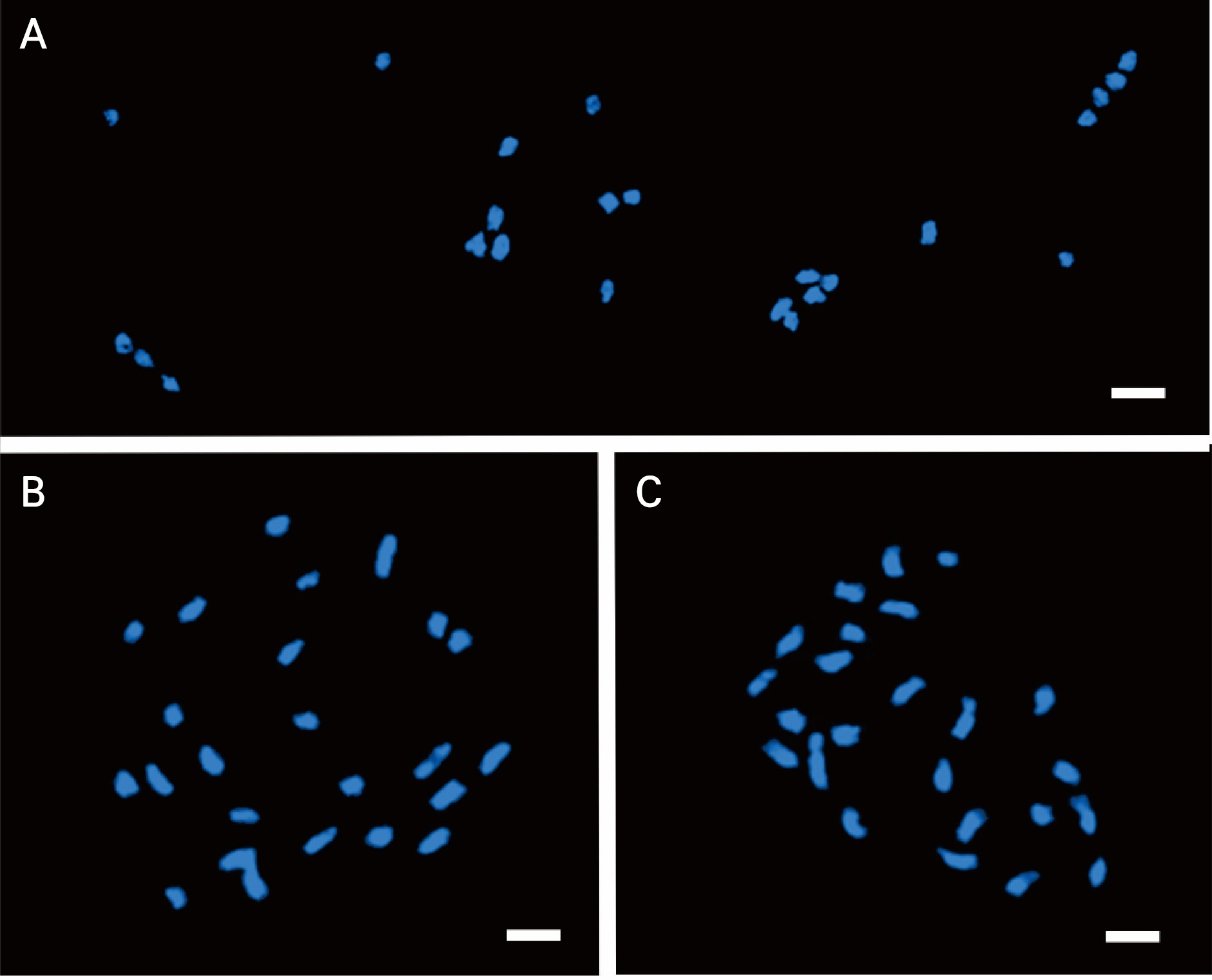
To verify the chromosome number, images of 20 adequately dispersed cells from each cultivar were acquired. This analysis identified 24 chromosomes in the cantaloupe, netted, and Hami melons, although their chromosome lengths varied (Fig. 2). In the somatic metaphase, the chromosome length of the cantaloupe type varied in the range of 1.72–2.8 µm (total length: 28.50 µm, total short arm length: 10.83 µm, total long arm length: 17.64 µm, Table 1). When compared with cantaloupe, the netted and Hami melons had higher total chromosome lengths 34.05 and 35.03 µm, respectively, (Tables 2 and 3).
Table 1.
Chromosome length and distribution in Cucumis melo var. cantalupensis ‘JCCM-02’as revealed by 5S and 18S rDNA FISH
| Chr. no | Chromosome length (µm) | Ribosomal DNA | Chr. Type | ||||
| Short arm | Long arm | Total | 5S | 18S | |||
| 1 | 1.17 ± 0.11z | 1.68 ± 0.22 | 2.85 ± 0.33 | - | - | ym | |
| 2 | 1.07 ± 0.08 | 1.73 ± 0.13 | 2.81 ± 0.21 | - | - | xsm | |
| 3 | 1.03 ± 0.03 | 1.63 ± 0.23 | 2.66 ± 0.26 | - | 2, S | m | |
| 4 | 1.02 ± 0.07 | 1.56 ± 0.14 | 2.58 ± 0.21 | - | - | m | |
| 5 | 1.01 ± 0.01 | 1.57 ± 0.23 | 2.58 ± 0.24 | - | - | m | |
| 6 | 0.93 ± 0.03 | 1.66 ± 0.09 | 2.59 ± 0.12 | 2, L | - | m | |
| 7 | 0.89 ± 0.05 | 1.33 ± 0.16 | 2.22 ± 0.21 | - | - | m | |
| 8 | 0.88 ± 0.11 | 1.67 ± 0.11 | 2.55 ± 0.22 | - | - | m | |
| 9 | 0.87 ± 0.11 | 1.24 ± 0.12 | 2.1 ± 0.23 | - | 2, S | m | |
| 10 | 0.83 ± 0.14 | 1.08 ± 0.11 | 1.9 ± 0.25 | - | - | m | |
| 11 | 0.62 ± 0.02 | 1.32 ± 0.20 | 1.94 ± 0.22 | - | - | sm | |
| 12 | 0.51 ± 0.03 | 1.17 ± 0.09 | 1.72 ± 0.12 | - | - | sm | |
| Total | 10.83 | 17.64 | 28.5 | ||||
Table 2.
Chromosome length and distribution in Cucumis melo var. reticulatus ‘Top Earl’s’, as revealed by 5S and 18S rDNA FISH
| Chr no. | Chromosome length (µm) | Ribosomal DNA | Chr. Type | ||||
| Short arm | Long arm | Total | 5S | 18S | |||
| 1 | 1.50 ± 0.01z | 1.89 ± 0.13 | 3.39 ± 0.14 | - | - | ym | |
| 2 | 1.27 ± 0.02 | 1.79 ± 0.17 | 3.06 ± 0.19 | - | - | m | |
| 3 | 1.19 ± 0.05 | 1.32 ± 0.06 | 2.51 ± 0.11 | - | 2, L | m | |
| 4 | 1.14 ± 0.01 | 2.02 ± 0.27 | 3.16 ± 0.28 | - | 2, L | m | |
| 5 | 1.13 ± 0.12 | 2.03 ± 0.10 | 3.16 ± 0.22 | - | - | xsm | |
| 6 | 1.03 ± 0.01 | 1.94 ± 0.07 | 2.97 ± 0.08 | - | - | sm | |
| 7 | 1.02 ± 0.04 | 1.4 ± 0.09 | 2.42 ± 0.13 | 2, S | - | m | |
| 8 | 1.01 ± 0.11 | 1.63 ± 0.37 | 2.63 ± 0.47 | - | - | m | |
| 9 | 0.89 ± 0.12 | 1.92 ± 0.15 | 2.81 ± 0.27 | - | - | sm | |
| 10 | 0.87 ± 0.02 | 1.99 ± 0.34 | 2.86 ± 0.36 | - | - | sm | |
| 11 | 0.86 ± 0.16 | 1.75 ± 0.41 | 2.61 ± 0.57 | - | - | sm | |
| 12 | 0.76 ± 0.11 | 1.75 ± 0.41 | 2.51 ± 0.51 | - | - | sm | |
| Total | 12.62 | 21.43 | 34.05 | ||||
5S and 18S ribosomal DNA sites
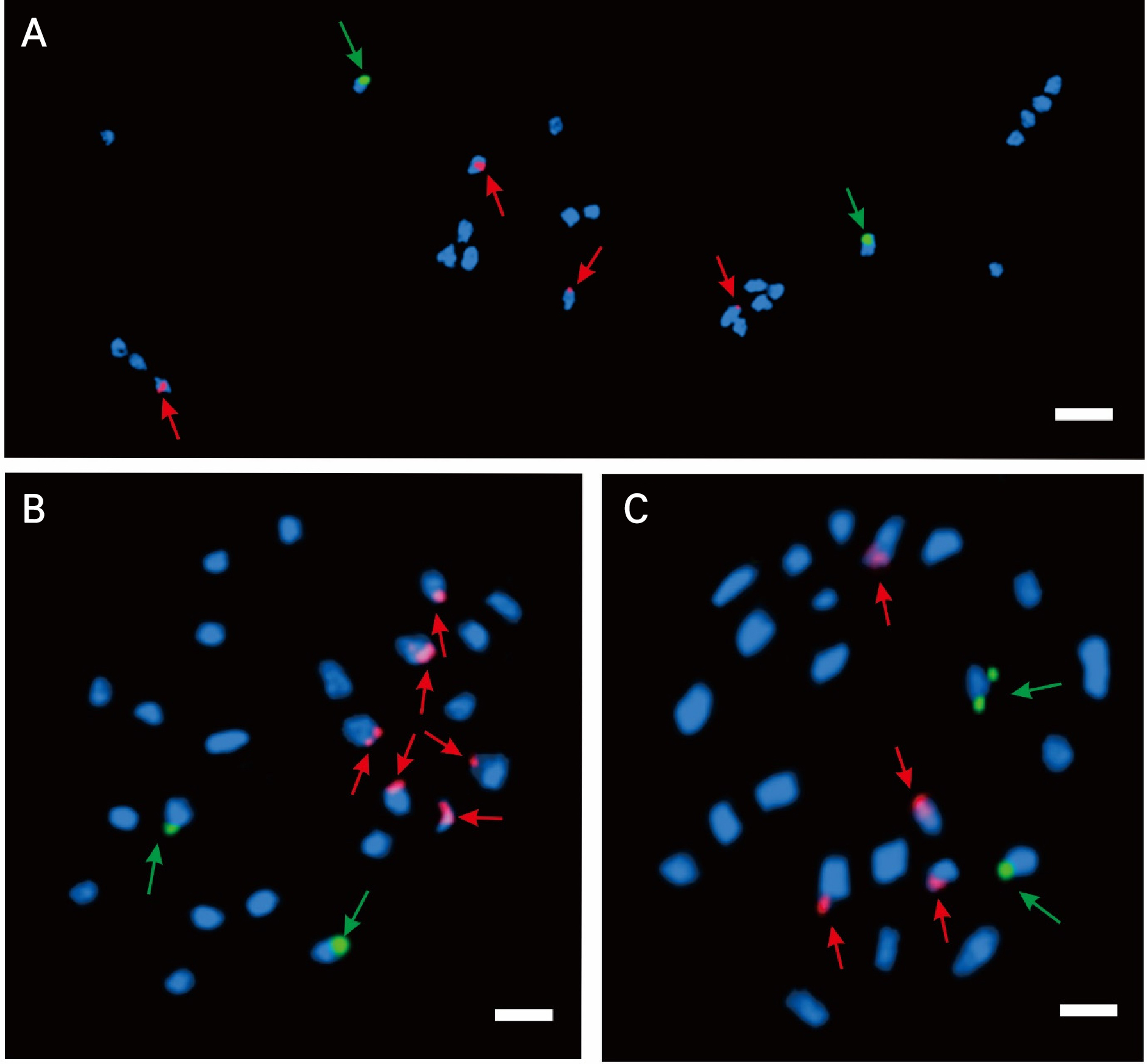
This study identified two 5S rDNA loci (green fluorescence) in the cantaloupe, Hami, and netted melons. Additionally, four 18S rDNA loci were observed in the cantaloupe and Hami melons. However, the netted melon had six 18S rDNA loci (Fig. 3). The 5S rDNA loci of the cantaloupe, netted, and Hami melons were found in specific regions of chromosomes 6, 7, and 10 (Fig. 4), respectively. However, the 18S rDNA loci exhibited distinct distribution patterns across several chromosomal regions. The cantaloupe had four rDNA signals in the short arms of chromosomes 3 and 10, the netted melon had four signals in the long arms of chromosomes 3 and 4 and two signals in the short arm of chromosome 11 (Fig. 4B), and the Hami melon had two signals in the long arm of chromosome 8 and the short arm of chromosome 11 (Fig. 4C).
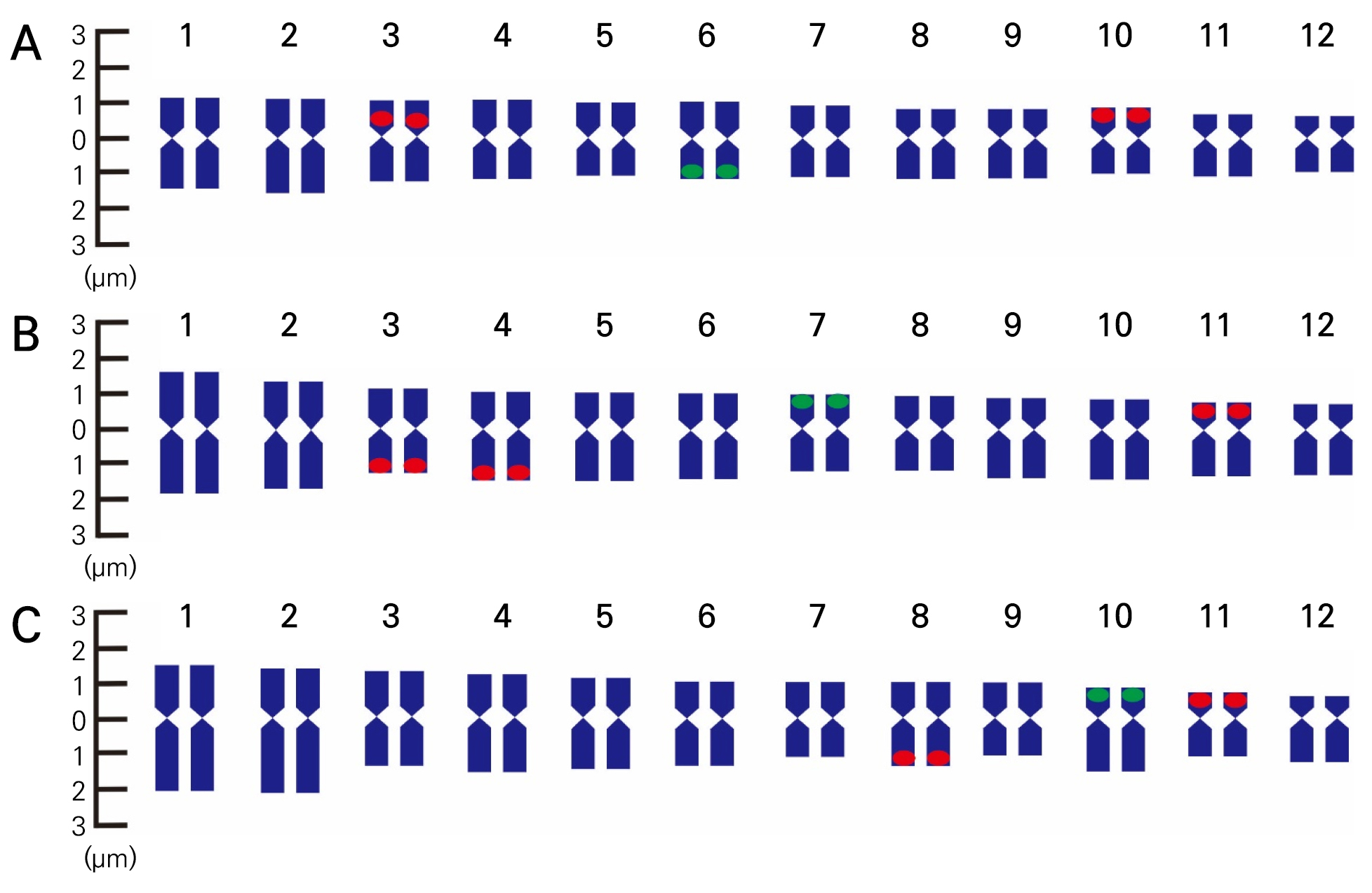
Fig. 4.
FISH karyotype analysis results of Cucumis melo var. cantalupensis ‘JCCM-02’ (A), C. melo var. reticulatus ‘Top Earl’s’ (B), and C. melo var. saccharinus ‘Hami Gold’ (C). Green and red represent the 5S rDNA and 18S rDNA probes, respectively. The chromosomes are arranged in descending order based on the short arm lengths.
Chromosome karyotyping
Based on the short arm length, the chromosomes were arranged in descending order. Depending on the centromere location, homologous chromosomes were categorized as metacentric or submetacentric pairs. Telocentric and subtelocentric chromosomes were not detected. The cantaloupe melon had 18 metacentric chromosomes and six submetacentric chromosomes, the netted melon had 12 metacentric and submetacentric chromosomes, and the Hami melon had 14 metacentric chromosomes and 10 submetacentric chromosomes (Tables 1, 2, 3).
Table 3.
Chromosome length and distribution in Cucumis melo var. saccharinus ‘Hami Gold’ as revealed by 5S and 18S rDNA FISH
| Chr no. | Chromosome length (µm) | Ribosomal DNA | Chr Type | ||||
| Short arm | Long arm | Total | 5S | 18S | |||
| 1 | 1.53 ± 0.12z | 2.16 ± 0.16 | 3.69 ± 0.28 | - | - | ym | |
| 2 | 1.42 ± 0.21 | 2.33 ± 0.27 | 3.75 ± 0.48 | - | - | xsm | |
| 3 | 1.34 ± 0.11 | 1.60 ± 0.25 | 2.94 ± 0.35 | - | - | m | |
| 4 | 1.28 ± 0.13 | 2.16 ± 0.41 | 3.44 ± 0.54 | - | - | m | |
| 5 | 1.26 ± 0.09 | 2.01 ± 0.13 | 3.27 ± 0.22 | - | - | m | |
| 6 | 1.16 ± 0.08 | 1.78 ± 0.41 | 2.94 ± 0.49 | - | - | m | |
| 7 | 1.08 ± 0.05 | 1.25 ± 0.54 | 2.33 ± 0.59 | - | - | m | |
| 8 | 1.02 ± 0.07 | 1.94 ± 0.16 | 2.96 ± 0.23 | - | 2, L | sm | |
| 9 | 1.01 ± 0.08 | 1.09 ± 0.20 | 2.09 ± 0.28 | - | - | m | |
| 10 | 0.93 ± 0.09 | 2.11 ± 0.32 | 3.04 ± 0.41 | 2, S | 2, L | sm | |
| 11 | 0.83 ± 0.06 | 1.27 ± 0.28 | 2.10 ± 0.34 | - | - | sm | |
| 12 | 0.69 ± 0.06 | 1.78 ± 0.46 | 2.47 ± 0.52 | - | - | sm | |
| Total | 13.54 | 21.48 | 35.02 | ||||
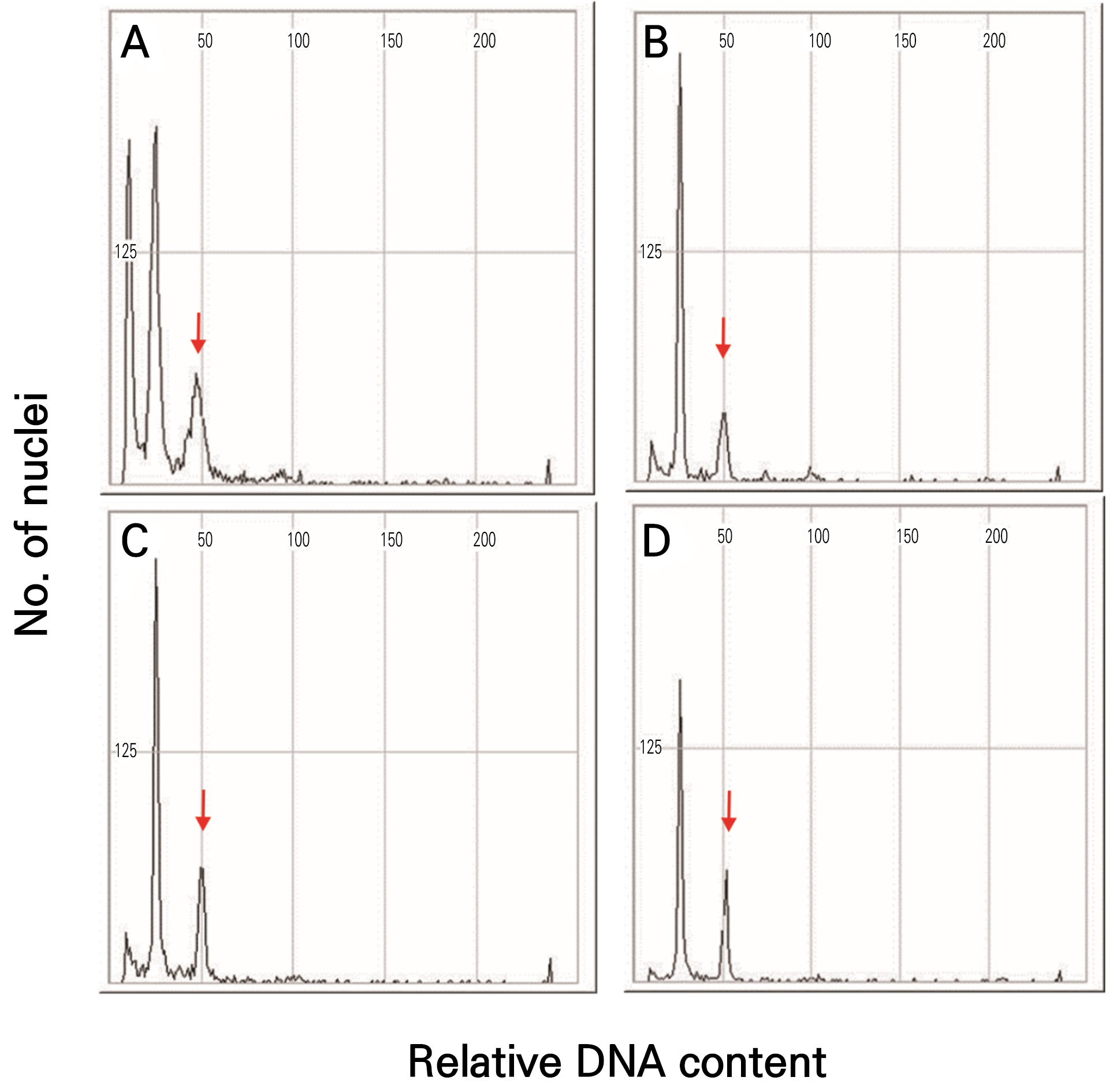
2C-DNA content
The histogram shows the peaks for the cantaloupe, netted, and Hami melons in channel 50 (Fig. 5). The three melon varieties had corresponding 2C-DNA contents of 1.15, 1.18, and 1.11 pg (Table 4). These results were obtained for the 2C genome, and the cantaloupe, netted, and Hami melons had contents of 1128.66, 1155.66, and 1084.03 Mbp, respectively (Table 4).
Table 4.
Putative DNA content and ploidy of the three melon varieties
Discussion
The identification of chromosomes is crucial in cytogenetic investigations. In melons, somatic metaphase chromosomes are small and similar in size, which makes them difficult to identify accurately (Liu et al., 2010). Chromosome number determination is crucial for cytogenetic research (Mohammad et al., 2020). Diploid melons have 12 sets of basic chromosomes (2n=24). However, the chromosome sizes varied across the varieties examined in this study, with the largest chromosome (2.85 µm) being more than twice as long as the smallest one (1.72 µm) in the cantaloupe melon. Chromosome size variations have previously been observed in C. melo var. inodorus (Karimzadeh et al., 2010), C. melo var. reticulatus (Karimzadeh et al., 2010), C. melo spp. melo Pang. (Ma et al., 1994), and C. melo var. Bartek (Daryono and Kumalawati, 2013). These variations are commonly associated with changes that occurred during the group’s evolution, such as genetic material removals or duplications (Auger and Sheridan, 2012).
FISH is a molecular cytogenetic tool that uses fluorescent-labeled probes to detect specific DNA sequences (Speicher and Carter, 2005), and it has been used to determine ploidy in plants (Waminal and Kim, 2015). 5S and 18S rDNA markers are extensively used in FISH, and the commonly utilized 5S rDNA loci consist of repeat 5S rRNA units together with intergeneric spacer regions. The 45S rRNA loci are made up of repeated 18S, 5.8S, and 25S rDNA units, as well as internal transcribed sequences (ITS1 and ITS2) and intergeneric spacer regions (Volkov et al., 2017). In this study, FISH revealed that cantaloupe and Hami melons have one pair of 5S rDNA loci and two pairs of 18S rDNA loci, whereas the netted melon has one pair of 5S rDNA loci and three pairs of 18S rDNA loci. A previous study of cotton found that most diploid plants possessed one pair of 5S rDNA loci (Gan et al., 2013), with a similar characteristic observed in the woody species of the Rubus genus (Wang et al., 2015). The 5S rDNA copy number, commonly assessed using FISH, is a reliable indicator of plant ploidy (Younis et al., 2015; Mohammad et al., 2020).
Karyotyping, a method of determining chromosomal variations and rDNA loci arrangements, also facilitates genetic map creation (Chung et al., 2018). The cytological tool described by Liang and Chen (2015) is widely used in empirical and theoretical studies. Because melon chromosomes are small and physically similar, it can be challenging to determine their karyotype. Based on our findings, the karyotype formulas for the cantaloupe, netted, and Hami melons were (9m + 3sm), (6m + 6m), and (7m + 5sm), respectively. The chromosomes were classified as metacentric or submetacentric depending on the centromere position. These results differ from observations of other melon varieties, such as C. melo var. Bartek (12m) (Daryono and Kumalawati, 2013), C. melo var. agrestis (2m + 9sm + 1st), C. melo var. ghurmi (3m + 8sm + 1st), C. melo var. momordica (3m + 9sm), and C. melo var. muskmelon. Karyotype formula variability suggests structural chromosomal changes, which may alter the centromere position owing to changes in the amount of DNA gained or lost between chromosomal arms (Stebbins, 1971). The Cucumis genus is made up of many species, and their karyotype variations are likely caused by centromere movement and other structural alterations (Han et al., 2009).
Flow cytometry allows rapid, precise, and straightforward assessments of DNA content for plant cytogenetic trait analysis (Galbraith, 2009). This method simplifies plant species classification in natural and agricultural contexts, enabling straightforward ploidy determination. Despite the availability of flow cytometric methods, C-values have been determined for only 2% of documented angiosperm species (Galbraith, 2009). This study revealed that cantaloupe, netted, and Hami melons have approximately equal 2C-DNA contents (1.15, 1.18, and 1.11 pg, respectively) and that the three melon varieties have comparable 2C genome sizes (1128.66, 1155.66, and 1084.03 Mbp, respectively).