Introduction
Materials and Methods
Plant Samples
Slide Preparation
Probe Preparation
Fluorescence in situ Hybridization (FISH)
Results
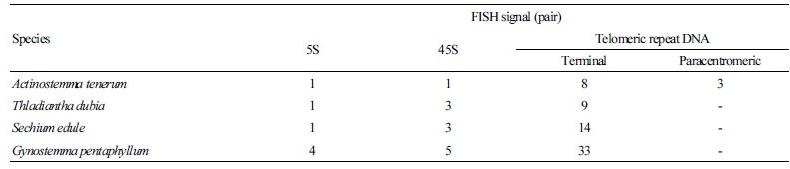
FISH with rDNA and Telomeric Repeats
FISH Karyotype
Discussion
Introduction
Cucurbitaceae is widely distributed throughout tropical and sub-tropical regions (Jobst et al., 1998). Numerous species in this family have been cultivated globally to accommodate human nutritional and economic needs (Ng, 1993; Kim et al., 2016), including cucumber, gourd, melon, watermelon, and pumpkins. These crops generate billions in revenue (Weng and Sun, 2012), and thus, there is considerable interest in breeding programs that aim to improve cucurbit quality.
Wild cucurbits also possess a variety of medicinal qualities that give them economic value. Medicinal Cucurbitaceae includes African cucumber (Momordica balsamina), wild cucumber (Cucurbita palmata), and wild melon (Lagenaria sphaerica) (Ntuli, 2007). Furthermore, some cucurbits such as Actinostemma lobatum and Sechium edule have antioxidative properties (Ordonez et al., 2006; Kim, 2010). In China, Thladiantha dubia and Gynostemma pentaphyllum are popular as ingredients for folk medicine. The pharmacological abilities attributed to these plants are extensive, including anti-analgesic, anti-inflammatory, and detoxification bioactivity (Razmovski-Naumovski et al., 2005; Tong et al., 2006; Tong et al., 2010; Zhang et al., 2010). Breeding programs incorporating wild cucurbits could generate improved crops with cucurbits’ pharmacological characteristics and other desirable traits, such as stress resistance (Anamthawat-Jónsson, 2001; Zamir, 2001). Indeed, many economically important plants (e.g., wheat, rice, rapeseed, some cucurbits) have benefitted from introgression with genetic material from wild varieties (Jiang et al., 1993; Brar and Khush, 1997; Humphreys et al., 1997; Snowdonet et al., 1997; Lashermes et al., 2000; Anamthawat-Jónsson, 2001). For instance, wild Cucumis hystrix was hybridized with domesticated Cucumis sativus to increase the genetic diversity of cucumber (Zhuang, 2006; Zhou et al., 2009; Delannay, 2010). Wild cucurbits like Cucurbita moschata (Formisano, 2010), Momordica charantia (Liao, 2012), and Citrullus spp. (Sain et al., 2002) have also been used to generate hybrids.
Molecular cytogenetic studies elucidate phylogenetic relationships (Heslop-Harrison, 1991; Jobst et al., 1998; Albert et al., 2010; Baloglu et al., 2015) and provide useful information for plant breeding (Garcia-Mas et al., 2012; Benjak et al., 2012; Guo et al., 2013), including polyploidy, physically mapping specific DNA sequences, and genome rearrangements (Khrustaleva and Kik, 2001; Devi et al., 2005; Kato et al., 2006; Macas et al., 2007; Huang et al., 2009; Sybenga, 2012). Knowing the closest cultivated relatives and chromosome characteristics of wild cucurbits would enhance their utility in breeding programs (Renner et al., 2007).
Fluorescence in situ hybridization (FISH) is a molecular technique used to map cytogenetic markers on chromosomes. It is useful for elucidating genome structure and identifying chromosomes (Leitch and Heslop-Harrison, 1992; Kato et al., 2005; Abd El-Twab and Kondo, 2006). With the use of rDNAs and known cytogenetic markers, studies have traced chromosomal rearrangement during evolution and clarified phylogenetic relationships (Devi et al., 2005; Coluccia et al., 2011; Miao et al., 2016). FISH has facilitated chromosomal characterization in wild cucurbits, allowing wild species to be further exploited in breeding programs (Leitch and Heslop-Harrison, 1992; Anamthawat-Jónsson, 2001; Xu et al., 2007; Koo et al., 2010; Sousa et al., 2012; Miao et al., 2016).
Here, we established triple-color FISH karyotypes of four Korean wild cucurbit species; Actinostemma tenerum Grift., Thladiantha dubia Bunge, Sechium edule (Jacq.) Swartz, and Gynostemma pentaphyllum, by visualizing the chromosomal distribution of three widely used cytogenetic markers: 5S and 45S rDNAs (ribosomal DNAs), as well as Arabidopsis-type telomeric repeats. Additionally, we discuss the role of polyploidy regarding the hexaploid cytotypes of G. pentaphyllum.
Materials and Methods
Plant Samples
The four Korean wild cucurbit species Actinostemma tenerum, Thladiantha dubia, Sechium edule, and Gynostemma pentaphyllum were collected near Moolangae park, Gwangju, Gyeonggi-Do, mountainside near Jeongseon, Gangwon-Do, near Sangwon Temple, Gangwon-Do, and the summit of Seowoobong orum, Hamdeok, Jeju-Do, Korea, respectively. Each species can be easily distinguished because it has distinct morphological characteristics (Lee, 2003). Fresh root tips were cut as ~2 cm length and pre-treated with 2 mM 8-hydroxyquinoline for 5 h at 18ºC before being fixed in Carnoy’s fixative for 2 h and stored in 70% ethanol.
Slide Preparation
Chromosome spreads were prepared following Waminal et al. (2011). Briefly, root tips were enzymatically digested with a pectolytic enzyme solution for 1 h. Protoplasts were resuspended in (9:1 v/v) aceto-ethanol. The suspension was then mounted on pre-warmed slides in a humidity chamber, air-dried, fixed in 2% formaldehyde for 5 min (Vrana et al., 2012), and dehydrated using an ethanol series treatment (70%, 90%, and 100%).
Probe Preparation
Following Matoba et al. (2007), 18S rDNA was PCR-amplified and labelled with DEAC-5-dUTP using nick-translation. Brassica oleracea 5S rDNA (Koo et al., 2002) was amplified and labelled through nick translation with Alexa Fluor® 488-5-dUTP (Invitrogen, Carlsbad, California, USA). The Arabidopsis-type telomere sequence was PCR-amplified (primers: 5'-TTTAGGG-3' and 5'-CCCTAAA-3') and labelled with Texas Red-5-dUTP (Perkin Elmer, NEL417001EA) following previous methods (Abd El-Twab and Kondo, 2006) with minor modifications.
Fluorescence in situ Hybridization (FISH)
The hybridization mixture comprised 50% formamide, 10% dextran sulfate, 2× Saline-sodium citrate buffer (SSC), 50 ng·uL-1 per DNA probe, and nuclease-free water. The mixture was denatured at 90ºC for 10 min and 40 µL was pipetted onto each slide. Chromosomes were denatured at 80ºC for 5 min and incubated overnight in a humidity chamber at 37ºC. Post-incubation stringency washes were performed: 2× SSC at 20ºC to 25ºC for 10 min, 0.1× SSC at 42°C for 25 min, and 2× SSC at RT for 5 min; followed by dehydration in an ethanol series (70%, 90%, 95%) at room temperature. Slides were air-dried and counterstained with 1 µg·mL 4', 6-diamidino-2-phenylindole (DAPI) in Vectashield (Vector Labs, H-1000, USA), then observed under an Olympus BX53 fluorescence microscope equipped with a Leica DFC365 FS CCD camera using an oil lens (×100 magnification). Captured images were processed using Cytovision ver. 7.2 (Leica Microsystems, Germany). Adobe Photoshop CS6 was used for image enhancement and ideogram preparation.
Results
FISH with rDNA and Telomeric Repeats
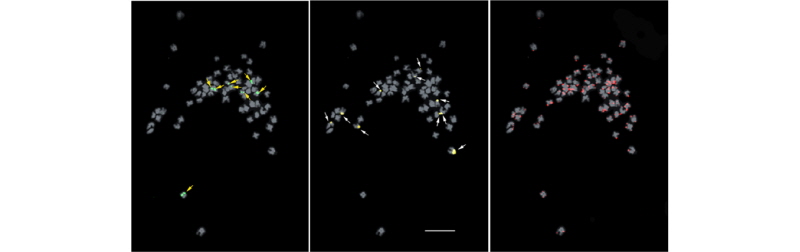
The A. tenerum genome contained one pair each of 5S and 45S rDNA signals (Fig. 1A). In both T. dubia and S.edule, one 5S pair and three 45S pairs were observed (Fig. 1B and 1C). Linkage of 5S and 45S rDNA was detected on chromosome 9 of
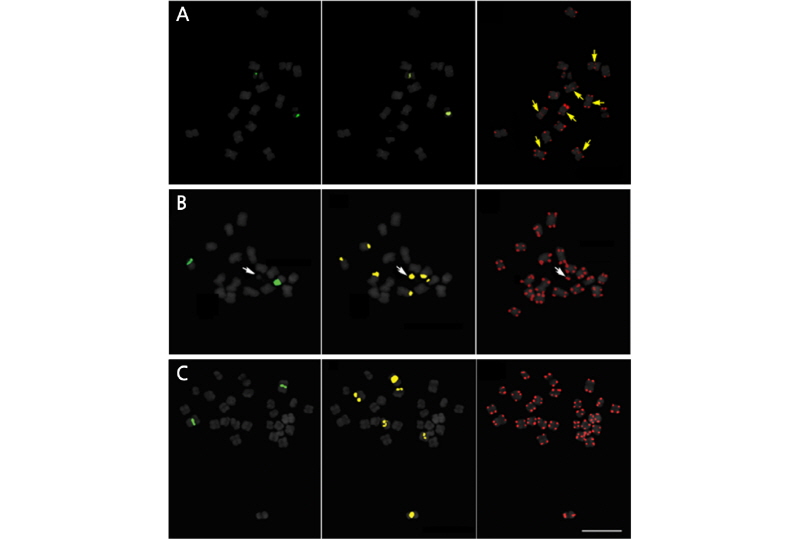
Fig. 1.
FISH signals of Actinostemma tenerum (A), Thladiantha dubia (B), Sechium edule (C) on somatic metaphase chromosomesusinga three-colorprobe cocktail. Figuresonthe left,middle andrightcolumnsshow5S rDNA,45SrDNA and telomere signals, respectively. White and yellow arrows indicate satellite chromosome and telomeric signals hybridized on the paracentromeric region, respectively. Bar = 10 μm.
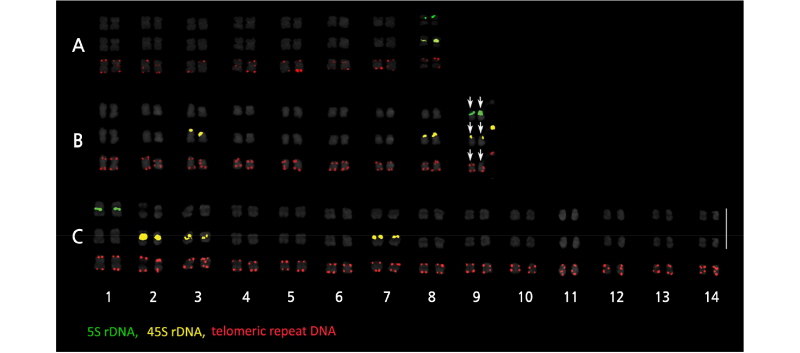
T. dubia (Fig. 1B), in contrast to the independent localization of rDNA repeats in S. edule (Fig. 1C). The hexaploid G. pentaphyllum exhibited more signals than the other three cucurbits, with four 5S loci and five 45S loci (Fig. 2). Table 2 summarizes the rDNA and telomeric signal distributions.
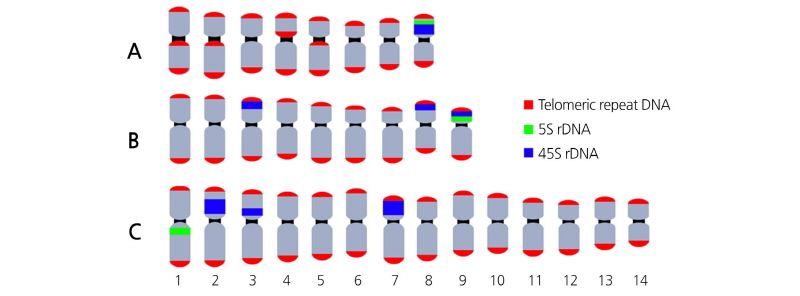
In A. tenerum, 5S rDNA was adjacent to 45S rDNA and hybridized on the short arm of chromosome 8 (Fig. 3A). In T. dubia, rDNA colocalization was observed on the short arm of chromosome 9, whereas two 45S rDNA individually localized on the terminal regions of chromosomes 3 and 8 (Fig. 3B). In S. edule, 5S rDNA localized to the paracentromeric region on the long arm of chromosome 1, while 45S rDNA localized to the interstitial regions on the short arm of chromosomes 2, 3, and 7, with signals in 2 and 7 being particularly intense (Fig. 3C). In G. pentaphyllum, 5S rDNA localized to paracentric or interstitial regions, whereas 45S rDNA localized to the terminal region (Fig. 2). In all four cucurbits, telomeric signals were present only on chromosomal termini, although an additional locus was detected on the paracentromeric region in A. tenerum.
FISH Karyotype
Chromosome number and arrangement of the four cucurbits were determined using FISH-based karyotype analysis with somatic metaphase chromosomes. The typical chromosome number of the family Cucurbitaceae is either 11 or 12 (Beevy and Kuriachan, 1996). However, some species are exceptions and possess 8, 9, and 14 chromosomes like in A. tenerum, T. dubia, and S. edule, respectively (Rice et al., 2015; Gao et al., 1995; De Wilde and Duyfjes, 2007; Tropicos.org, 2017). Chromosomes were paired according to probe signals and arranged in descending order of length (Fig. 3). Corresponding karyotypic ideograms are shown in Fig. 4.
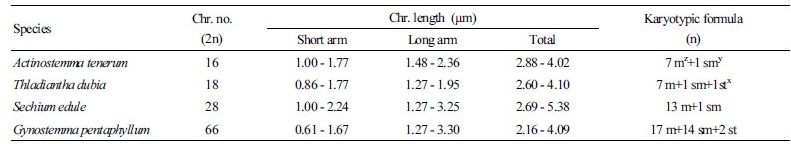
The chromosome complement of A. tenerum was 2n = 2x = 16 (Fig. 3A), with chromosome lengths of 2.88-4.02 µm that comprised seven metacentric and one submetacentric chromosomes (Table 1). That of S. edule was 2n = 2x = 28 (Fig. 3B), with lengths of 2.69 - 5.38 µm comprising 13 metacentric and one submetacentric chromosomes (Table 1). That of T. dubia was 2n = 2x = 18 (Fig. 3C), with seven metacentric, one submetacentric, and one subtelocentric chromosomes around 2.60 - 4.10 µm in length. Finally, that of G. pentaphyllum was 2n = 6x = 66, but the small size of the chromosomes precluded length measurements and karyotype analysis.
Discussion
Most Cucurbitaceae species have a basic chromosome number of x = 12 (Whitaker, 1933; Schaefer and Renner, 2010), but the putative basic chromosome number of ancestral cucurbits is x = 14. Other variations were thought to have evolved either via aneuploid reduction or amplification (Beevy and Kuriachan, 1996). The chromosome number variation between cucumber (Cucums sativus, x = 7) and melon (Cucumis melo, x = 12) provide a clear example of this genomic amplification or downsizing through chromosomal fragmentation or fusion (Ramachandran and Seshadri, 1986; Koo et al., 2010).
Likewise, the four species in this study varied in chromosome numbers, with haploid counts of 8, 9, 11, and 14 for A. tenerum,
T. dubia, G. pentaphyllum, and S. edule, respectively (Table 1). In addition, these results support previous reports on their chromosome numbers (Fay, 2011; Rice et al., 2015). These patterns reflect general observations of chromosome variation in the Cucurbitaceae family (Whitaker, 1933; Jeffrey, 1967; Jeffrey, 1980).
Mobile rDNA clusters have been described in a number of plants (Raskin et al., 2008) and verified through observed variation in rDNA localization and number between closely related species, or even within a species (Castilho and Heslop-Harrison, 1995; Li and Arumuganathan, 2001). Frequently, rDNAs localizing on separate chromosomes lead to more efficient gene conversion, unequal crossing-over without disruptive interference, and rDNA repeat duplications or deletions (Martins and Galetti Jr, 1999; Martins and Galetti Jr, 2001; Koo et al., 2010; Liu and Davis, 2011). On the other hand, interstitial localization of 5S rDNA loci appears to prevent the dispersion of this repeat during chromosomal rearrangement, whereas 45S rDNA residing on distal or terminal regions are more prone to genomic dispersion (Waminal and Kim, 2012). Thus, transposition and dispersion of 45S rDNA are favored over 5S rDNA. In support of this tendency, we found more 45S rDNA loci than 5S rDNA loci across the four cucurbits. Another remarkable pattern observed was rDNA loci colocalization in T. dubia, implying a syntenic region carrying a shared ancestral trait (Bertioli et al., 2009; Marquioni et al., 2013). In addition, the unusual interstitial localization of telomeric repeats in A. tenerum may be artifacts of chromosome fusion during speciation from an ancestral genome (Fuchs et al., 1995; Datson and Murray, 2006; Książczyk et al., 2010). This observation is likely, considering the lesser basechromosome number in Actinostemma compared with the Cucurbitaceaeconsensus of x = 11 or 12, much like the fusion in Cucumis sativus at the divergence from C. melo (Koo et al., 2010).
The hexaploid cytotypes of G. pentaphyllum revealed rDNA numbers that are disproportionate to its ploidy level, suggesting that rDNA might have been eliminated during polyploidization (Mishima et al., 2002; Balao et al., 2011). Such reduction is tolerable because the initial number of rDNA sites exceeded the amount sufficient for sustaining normal cellular activity (Liu and Davis, 2011). Polyploidization is considered to be highly influential in angiosperm evolution (Cota and Philbrick, 1994; Ntuli, 2007). Among Cucurbitaceae, polyploidization occurred in numerous genera,particularly Gynostemma (Agarwal and Roy, 1976). Thefrequency of polyploidization inthis genusimplies a key rolein species diversification (Singh, 1979; Beevy and Kuriachan, 1996), beginning from a basic chromosome number of x = 11 in Gynostemma to many other varieties (2n = 22, 44, 66, 88) (Gao et al., 1995; De Wilde and Duyfjes, 2007). Notably, G. pentaphyllum ploidy is thought to be derived from autopolyplodization (Jiang et al., 2009; Li et al., 2012), wherein a single species exhibits homologous chromosome doubling (Parisod et al., 2010).
Phenotypic shifts are correlated with polyploidization (Balao et al., 2011) and could lead to trait modification in a species (Sax and Sax, 1937; Ntuli, 2007). For instance, genome duplication facilitated the distributional expansion of Achillea borealis, a polyploid species compared to its diploid cytotype, by enhancing adaptation to a wider environmental variation (Ramsey, 2011). Furthermore, the emergence of polyploidization may enhance a species’ physiological mechanisms and increase tolerance against environmental stressors, such as drought and temperature fluctuations by gene redundancy (Li et al., 1996; Schulze et al., 2005). This effect explains how polyploidization in G. pentaphyllum could contribute to the expansion of its geographical range and outcompeting other Gynostemma (Jiang et al., 2009). Furthermore, G. pentaphyllum has several medical functions that appear to be enhanced in accordance with its ploidy level (Liao et al., 2011; Niu et al., 2013). Specifically, medicinal properties were stronger in the polyploid than in the diploid G. pentaphyllum (Meyers and Levin, 2006; Otto, 2007).
In conclusion, our study describes triple-color FISH karyotypes of the four Korean wild cucurbit species, which will be useful in cucurbit breeding programs and for elucidating their genome structure. Clarifying plant genome structures allows the establishment of accurate phylogenies that can be used to trace wild relatives with desirable agronomic and medicinal traits to be incorporated into currently cultivated cucurbits (Devi., 2005; Balao et al., 2011; Coluccia et al., 2011; Miao et al., 2016). Future studies can refine our understanding of Cucurbitaceae genome constitution with the use of techniques such as self-genomic in situ hybridization or genome-wide analysis of next-generation sequencing data to generate repeat-based probes as cytogenetic markers.