Introduction
Materials and Methods
Plant Materials
Production of Homozygous S1 Seedlings of ‘Hirado Buntan’ for the S Gene
Pollination of Citrus Accessions by Homozygous S1 Seedlings (S9S9 or S10S10)
Variation of the Number of Pollen Tubes in Lower Styles of Back-cross-incompatible Pistils (S9S10) When Pollinated with ‘Hirado Buntan’ (S9S10)
Variation of the Number of Pollen Tubes in the Lower Styles of Incompatible Pistils of Citrus Plants Carrying S9 and/or S10 Alleles When Pollinated with S9 or S10 Pollen
Results
Determination of Citrus Accessions with S9 Pollen
Determination of Citrus Accessions with S10 Pollen
Variation of the Number of Pollen Tubes in Lower Styles of Back-cross-incompatible Pistils (S9S10) Pollinated with ‘Hirado Buntan’ (S9S10)
Variation of the Number of Pollen Tubes in the Lower Styles of Incompatible Pistils of Citrus Plants Carrying S9 and/or S10 Alleles When Pollinated with S9 or S10 Pollen
Discussion
Distribution of Citrus Accessions with S9 and/or S10 Alleles
Characteristics of the S Allele Frequency for S9 and S10 Alleles
Differences in the Self-incompatibility Reaction in Lower Styles between S9 and S10 Alleles
Conclusion
Introduction
Citrus is cultivated throughout the world, from tropical to temperate climates where there are suitable soils, sufficient moisture, and optimal temperatures (Soost and Roose, 1996). The importance of the citrus industry to agriculture and the economy is demonstrated by this large-scale production. Citrus is known to have gametophytic self-incompatibility (SI) governed by a single gene (S) in pummelo [Citrus maxima (Burm.) Merr.], mandarin (Citrus spp.), and their hybrids (Soost, 1987; Yamamoto et al., 2006; Kim et al., 2011; Ngo et al., 2019). SI is an important trait for breeding and seedless fruit production in Citrus cultivars with a high degree of parthenocarpy (Iwamasa and Oba, 1980; Vardi et al., 2000). For Citrus, however, there is very limited information about the self-incompatibility S gene sequence (Honsho et al., 2019; Liang et al., 2020), S allele variation, S allele frequencies, and the S genotypes of cultivars (Kim et al., 2020).
Ngo et al. (2019) studied the patterns and strength of self-incompatibility in 121 Citrus accessions, including 77 self-incompatible pummelos. They found that the degree of SI in the stigmas, upper styles, middle styles, and basal styles of the self-incompatible pistils differed and could be divided into three categories: high, moderate, and low. Eventually, there were no or only a few pollen tubes in the base of the self-pollinated styles of these self-incompatible accessions. These results suggested that self-incompatibility in a given Citrus accession could be determined only by observation of the pollen tubes in the basal style (Ngo et al., 2019).
To elucidate the self-incompatibility genotypes of Citrus cultivars, Ngo et al. (2011) used Got-3 allozyme markers associated with the self-incompatibility S gene and proposed the S1S2 genotype for ‘Banpeiyu’ (Citrus maxima), S1S3 for ‘Tosa Buntan’ (Citrus maxima), S4S5 for ‘Hassaku’ (Citrus hassaku), S6S7 for ‘Yuge Hyokan’ (Citrus yuge-hyokan), and S1S8 for ‘Hyuganatsu’ (Citrus tamurana). For the S1 and S2 alleles proposed by Ngo et al. (2011), Kim et al. (2011) determined the S genotypes in seven full- or semi-self-incompatible Citrus accessions by pollination with two homozygous S1 seedlings (S1S1 and S2S2), each derived from self-pollination of the self-incompatible ‘Banpeiyu’ (S1S2) pummelo. For the S4 and S5 alleles proposed by Ngo et al. (2011), Zhou et al. (2018) determined 12 full- and semi-self-incompatibility S genotypes and one full-self-compatibility S genotype (SfSf) in Citrus accessions by pollination with two homozygous S1 seedlings (S4S4 and S5S5), each from self-pollination of the self-incompatible ‘Hassaku’ (S4S5). For the S3 allele proposed by Ngo et al. (2011) and the S11 allele discovered by Kim et al. (2020), Kim et al. (2020) determined the S genotypes in 28 full- or semi-self-incompatible Citrus accessions by pollination with two homozygous S1 seedlings (S3S3 and S11S11), each from self-pollination of the self-incompatible Clementine (S3S11). In these S-genotyping studies with more than 100 full- and semi-self-incompatible Citrus accessions, the S allele frequency was calculated for S1 and S2 (Kim et al., 2011), S3 (Kim et al., 2020), S4and S5 (Zhou et al., 2018), and S11 (Kim et al., 2020) alleles.
In the previous study for S9 and S10alleles (Kim et al., 2010), homozygous S1 seedlings (SaSa and SbSb) of ‘Hirado Buntan’ (SaSb) were produced through bud self-pollination and allelic symbols of S9 and S10 were determined, respectively, for Sa and Sb, and then using the S9 and S10alleles, pollen detection of Citrus accessions with S9 and S10 alleles was carried out focusing on a small number of pummelos growing on Hirado Island adjacent to the northernmost part of Kyushu Island, Japan. A total of only 18 Citrus accessions was used for the determination and detection of S9and S10alleles with the homozygous S1 seedlings (S9S9 and S10S10) of ‘Hirado Buntan’ in the previous study. Because of the small number of accessions, we could not determine the origin of the S9 and S10 alleles, the rates of Citrus accessions with S9 or S10 alleles, and S9 and S10 allele frequencies in pummelo (a source of S alleles) and Citrus accessions (introgression of S alleles from pummelo). The present study was carried out to clarify the origin of the S9 and S10 alleles and to calculate the rates of Citrus accessions and pummelos with S9 or S10 alleles and the S9 and S10 allele frequencies in pummelo and Citrus accessions through S-genotyping of 146 Citrus accessions (not including artificial hybrid cultivars) with S9 or S10 alleles. When comparing the results of S-genotyping between pummelo (source of S alleles) and Citrus accessions (introgression of S alleles from pummelo), the genetic diversity in pummelo and its relation to the development and evolution of full-self-incompatible and semi-self-incompatible Citrus species and cultivars determined using S alleles as markers. Here, we aimed to uncover the genetic diversity in pummelo and the development and evolution of Citrus species and cultivars by monitoring their S alleles.
Materials and Methods
Plant Materials
We collected 146 Citrus accessions, including 82 pummelo accessions, to use for pollination with two homozygous S1 seedlings (S9S9 and S10S10) of ‘Hirado Buntan’ [Citrus maxima (Burm.) Merr.]. Most Citrus cultivars included in these accessions were provided by the National Institute of Fruit Tree Science (NIFTS), the Kumamoto Prefectural Fruit Tree Experiment Station (KMPFTES), and the Kagoshima Prefectural Fruit Tree Experiment Station (KSPFTES). All Citrus accessions and homozygous S1 seedlings (S9S9 and S10S10) were trees more than ten years old, grown in the Experimental Farm of Kyushu University, Sasaguri, Fukuoka. Self-incompatibility in these accessions had been determined through the observation of pollen tube arrest in their style base eight days after self-pollination (Ngo et al., 2019).
Production of Homozygous S1 Seedlings of ‘Hirado Buntan’ for the S Gene
Self-incompatible ‘Hirado Buntan’ was used to produce S1 seedlings homozygous for the S gene (Wakana et al., 2004). Seeds were collected and planted from fruits obtained by bud pollination on ‘Hirado Buntan’, and these S1 seedlings were top-worked on twenty-year-old ‘Hashimoto Wase’ satsuma mandarin (Citrus unshiu) trees and allowed to grow for more than ten years, until their flowers were available for pollination examination. The S genotype of the top-worked S1 seedlings homozygous for the S gene (S9S9 or S10S10) was determined by Kim et al. (2010). Of the S1 seedlings homozygous for the S gene, two S1 seedlings (HBS3 and HBS4) with the S9S9 genotype and two S1 seedlings (Hirado Buntan Seedling No.2; HBS2 and Hirado Buntan Seedling No.11; HBS11) with the S10S10 genotype were used to pollinate Citrus accessions. In addition to the four S1 seedlings, HBS19 and HBS25 seedlings with the S9S9 genotype and an HBS20 seedling with the S10S10 genotype, which were determined in the present study by a self-incompatibility reaction in the lower styles eight days after pollination with S9 and S10 pollen of HBS3 and HBS2, respectively, were used for the present study because they produced enough pollen to carry out the pollination experiment.
Pollination of Citrus Accessions by Homozygous S1 Seedlings (S9S9 or S10S10)
To examine whether the 146 Citrus accessions share one or both of the S alleles (S9 and/or S10) with ‘Hirado Buntan’ (S9S10), they were pollinated with the homozygous S1 seedlings of ‘Hirado Buntan’, i.e., HBS3, HBS4, HBS19, and HBS25 seedlings with the S9S9 genotype and HBS2, HBS11, and HBS20 seedlings with the S10S10 genotype. Normal flowers from which petals and stamens were removed just before flowering were selected, and the stigmas of the normal pistils were pollinated with the S9 or S10 pollen of ‘Hirado Buntan’ homozygous S1 seedlings until they showed a yellow color; they were then bagged to prevent further pollination. Five emasculated flowers were pollinated per Citrus accession in each year. Eight days after pollination, the styles were collected and fixed with acetic acid:alcohol (1:3 v/v). These were stored in 70% ethanol at 4 ℃ until use and were softened with 0.8 N sodium hydroxide solution for 24 hours when used. After that, the softened styles were stained overnight with a solution of 0.1% aniline blue. The stained pistils were divided into five parts (stigma, upper style, middle style, lower style, and ovary). The lower styles were observed under a fluorescence microscope (Nikon 50i). When no or fewer than ten pollen tubes were observed in the lower styles, their upper styles or stigmas were observed to ascertain the presence of a large number of pollen tubes. The pollen-tube observation and the determination of S alleles were carried out according to the report of Zhou et al. (2018). The pollen-tube observation was carried out using three pistils per accession and was repeated for two to four years, especially in the accessions in which the incompatibility reaction is not clear. The mean number of pollen tubes in the lower styles observed in each year was averaged per year in each accession.
In the previous study, we determined the S genotype of ‘Kabusu’ sour orange (SfS9) by pollination with a homozygous S1 seedling (S9S9) of ‘Hirado Buntan’ (Kim et al., 2010). However, further certification of the ‘Kabusu’ S genotype was carried out in the present study, since it was determined using two pistils and since ‘Zadaidai’ sour orange had a different S genotype (SfS11) (Kim et al., 2020) from ‘Kabusu’ despite that they are considered as clones (personal communication).
Variation of the Number of Pollen Tubes in Lower Styles of Back-cross-incompatible Pistils (S9S10) When Pollinated with ‘Hirado Buntan’ (S9S10)
Five heterozygous S1 seedlings (HBS5, HBS15, HBS18, HBS25 and HBS27) with the S9S10 genotype were pollinated with ‘Hirado Buntan’. The number of pollen tubes was examined in lower styles. It was also examined in their upper styles to check the existence of many pollen tubes. The procedure of pollination and pollen tube observation was similar to that mentioned above.
Variation of the Number of Pollen Tubes in the Lower Styles of Incompatible Pistils of Citrus Plants Carrying S9 and/or S10 Alleles When Pollinated with S9 or S10 Pollen
In the present study, when ‘Hirado Buntan’ was pollinated with S9 pollen of ‘Hirado Buntan’ S1 seedlings, almost no S9 pollen tubes were observed in the lower styles, whereas when pollinated with S10 pollen of another ‘Hirado Buntan’ S1 seedling, nine S9 pollen tubes on average were observed per lower style. To examine the difference in the degree of SI in the lower part of styles between S9 and S10 pollen tubes, seven S1 seedlings [HBS2 (S10S10), HBS3 (S9S9), HBS4 (S9S9), HBS7 (S9S9), HBS18 (S9S10), HBS19 (S9S9) and HBS25 (S9S9)] and six Citrus accessions [‘Hirado Buntan’ (S9S10), ‘Kabusu’ (SfS9), Amakusa No.6 (S9S10), Yatsushiro No.4 (S10S11) , ‘Bangkok Buntan’ (S10S?) and Kawanabe No.2 (S5S10) ] carrying S9 and/or S10 alleles were used. HBS3 (S9S9), HBS4 (S9S9) and HBS25 (S9S9) were used as pollen parents generating S9 pollen, while HBS2 (S10S10) was used as a pollen parent generating S10 pollen. The number of pollen tubes was examined in lower styles, and also in their upper styles to check the existence of many pollen tubes. The procedure of pollination with S9 or S10 pollen and pollen tube observation was similar to that mentioned above.
Results
Determination of Citrus Accessions with S9 Pollen
When the S9 pollen of ‘Hirado Buntan’ pummelo homozygous S1 seedlings (S9S9) was pollinated to 126 Citrus accessions, many S9 pollen tubes penetrated into the lower styles of 122 accessions (Table 1 and Fig. 1). The average number of pollen tubes observed in the lower styles of the 122 accessions ranged from 38.0 to 488.7. This result suggested that these accessions show cross compatibility with S9 pollen and do not have an S9 allele. In four of the 126 accessions, no or only a few pollen tubes penetrated into the lower styles: 0.5 for the Amakusa No. 6 pummelo, 1.0 for the ‘Hirado Buntan’ pummelo, 0.0 for the ‘Kabusu’ sour orange, and 0.0 for the ‘Kikudaidai’ sour orange relative. This result suggested that these accessions show cross incompatibility with S9 pollen and have an S9 allele.
Table 1.
Determination of Citrus accessions with S9 and/or S10 alleles using pollen of homozygous S1 seedlings of ‘Hirado Buntan’ (S9S10) for the self-incompatibility (S) gene
|
Scientific name, plant No., accession (Tanaka’s classification No.; Tanaka, 1969)z, and origin | S9 polleny | S10 polleny |
Presence (+) or absence (‑) of S9 and S10 alleles |
Determined S genotypex | |||
|
No. of flowers pollinated |
Mean No. of pollen tubes in style base |
No. of flowers pollinated |
Mean No. of pollen tubes in style base | ||||
| Lime, citron and relatives | |||||||
| C. limon (L.) Burm.f. (36) | |||||||
| 1. Allen Eureka*? | 3 | 146.0 | 3 | 109.0 | ‑ S9‑ S10 | SfS? | |
| 2. Cook Eureka*? | 3 | 74.3 | 3 | 179.7 | ‑ S9‑ S10 | SfS? | |
| 3. Lisbon*? | 3 | 228.3 | 3 | 224.3 | ‑ S9‑ S10 | SfS? | |
| 4. Villafranca*? | 3 | 213.0 | 3 | 177.0 | ‑ S9‑ S10 | SfS? | |
| Pummelo and relatives | |||||||
| C. maxima (Burm.) Merr. (56) | |||||||
| 5. Akune No.1** Japan | 3 | 158.7 | new | - | ‑ S9 ne | S?S? | |
| 6. Akune No.4** Japan | ne | - | 3 | 266.3 | ne ‑ S10 | S?S? | |
| 7. Akune No.6** Japan | 2 | 180.5 | 6 | 127.7 | ‑ S9‑ S10 | S?S? | |
| 8. Amakusa No.1** Japan | 3 | 176.3 | 3 | 120.7 | ‑ S9‑ S10 | S11S? | |
| 9. Amakusa No.2** Japan | 3 | 161.3 | 3 | 98.3 | ‑ S9‑ S10 | S5S? | |
| 10. Amakusa No.3** Japan | 3 | 337.7 | 3 | 186.7 | ‑ S9‑ S10 | S?S? | |
| 11. Amakusa No.4** Japan | 3 | 145.3 | 3 | 254.7 | ‑ S9‑ S10 | S?S? | |
| 12. Amakusa No.6** Japan | 6 | 0.5 | 6 | 0.0 | +S9 +S10 | S9S10 | |
| 13. Amakusa No.8** Japan | 5 | 117.6 | 3 | 382.7 | ‑ S9‑ S10 | S?S? | |
| 14. Amakusa No.12** Japan | 3 | 239.7 | 3 | 239.0 | ‑ S9‑ S10 | S?S? | |
| 15. Amakusa No.13** Japan | 3 | 122.3 | ne | - | ‑ S9 ne | S11S? | |
| 16. Amakusa No.14** Japan | 6 | 294.8 | 3 | 298.7 | ‑ S9‑ S10 | S?S? | |
| 17. Amakusa No.15** Japan | 6 | 294.5 | 3 | 243.7 | ‑ S9‑ S10 | S?S? | |
| 18. Amakusa No.17** Japan | 3 | 185.0 | 3 | 183.3 | ‑ S9‑ S10 | S11S? | |
| 19. Amami No.1** Japan | 3 | 223.3 | 6 | 208.3 | ‑ S9‑ S10 | S?S? | |
| 20. Amami No.2** Japan | 5 | 176.2 | 4 | 128.7 | ‑ S9‑ S10 | S?S? | |
| 21. Amami No.3** Japan | 6 | 238.2 | 4 | 136.5 | ‑ S9‑ S10 | S2S3 | |
| 22. Amami No.5** Japan | ne | ‑ | 4 | 149.6 | ne ‑ S10 | S?S? | |
| 23. Amami No.6** Japan | 4 | 159.0 | 3 | 264.3 | ‑ S9‑ S10 | S?S? | |
| 24. Amami No.7** Japan | 3 | 292.0 | ne | - | ‑ S9 ne | S3S? | |
| 25. Bangladesh No.48** Bangladesh | 2 | 278.0 | 2 | 166.0 | ‑ S9‑ S10 | S?S? | |
| 26. Bangladesh No.49** Bangladesh | 6 | 220.2 | 4 | 160.5 | ‑ S9‑ S10 | S?S? | |
| 27. Bangladesh No.52** Bangladesh | 6 | 261.0 | 4 | 107.2 | ‑ S9‑ S10 | S3S? | |
| 28. Bangkok Buntan (KPFTES)**Thai | 3 | 150.0 | 3 | 0.0 | ‑ S9 +S10 | S10S? | |
| 29. Banpeiyu**Vietnam | 3 | 120.7 | 3 | 170.3 | ‑ S9‑ S10 | S1S2 | |
| 30. Bansei Siam Buntan**Thai | 6 | 176.9 | 6 | 241.0 | ‑ S9‑ S10 | S11S? | |
| 31. Chandler**USA | 5 | 367.0 | 8 | 229.5 | ‑ S9‑ S10 | S4S11 | |
| 32. Hirado Buntan**Japan | 6 | 1.0 | 3 | 9.0 | +S9 +S10 | S9S10 | |
| 33. Hirado No.1** Japan | ne | - | 6 | 171.5 | ‑ S9‑ S10 | S?S? | |
| 34. Hirado No.2** Japan | ne | - | 3 | 192.3 | ‑ S9‑ S10 | S?S? | |
| 35. Hirado No.3** Japan | 6 | 188.3 | 4 | 225.9 | ‑ S9‑ S10 | S?S? | |
| 36. Hirado No.4** Japan | 3 | 214.0 | 6 | 238.0 | ‑ S9‑ S10 | S?S? | |
| 37. Hirado No.6** Japan | 3 | 113.0 | 3 | 141.7 | ‑ S9‑ S10 | S?S? | |
| 38. Hirado No.12** Japan | 3 | 317.7 | 5 | 262.6 | ‑ S9‑ S10 | S?S? | |
| 39. Hirado No.13** Japan | 3 | 377.7 | 3 | 299.0 | ‑ S9‑ S10 | S?S? | |
| 40. Hirado No.14** Japan | 3 | 201.7 | 3 | 209.7 | ‑ S9‑ S10 | S?S? | |
| 41. Honda Buntan** Japan | 3 | 229.0 | ne | - | ‑ S9 ne | S?S? | |
| 42. Ibusuki Buntan** Japan | ne | - | 3 | 77.3 | ne ‑ S10 | S1S? | |
| 43. Indonesia No.2050** Indonesia | 6 | 305.7 | 3 | 202.7 | ‑ S9‑ S10 | S?S? | |
| 44. Indonesia No.2080** Indonesia | 3 | 289.7 | 3 | 239.6 | ‑ S9‑ S10 | S1S11 | |
| 45. Ipoh No.1**Malaysia | 2 | 241.0 | 3 | 181.0 | ‑ S9‑ S10 | S2S? | |
| 46. Ipoh No.3** Malaysia | 6 | 309.7 | 4 | 233.7 | ‑ S9‑ S10 | S2S11 | |
| 47. Ipoh No.5** Malaysia | 4 | 131.2 | 6 | 134.4 | ‑ S9‑ S10 | S2S? | |
| 48. Iriki Buntan** Japan | 4 | 274.3 | 3 | 326.3 | ‑ S9‑ S10 | S1S2 | |
| 49. Kaopan** Thai | 6 | 391.5 | 6 | 182.0 | ‑ S9‑ S10 | S1S2 | |
| 50. Kaophuang** Thai | 3 | 194.0 | 3 | 264.7 | ‑ S9‑ S10 | S?S? | |
| 51. Kawanabe Buntan** Japan | 3 | 299.0 | 3 | 357.7 | ‑ S9‑ S10 | S?S? | |
| 52. Kawanabe No.2** Japan | 3 | 153.7 | 3 | 1.7 | ‑ S9 +S10 | S5S10 | |
| 53. Kirapeiyu**Taiwan | 3 | 356.3 | 3 | 163.3 | ‑ S9‑ S10 | S1S? | |
| 54. Koshiki No.1** Japan | 3 | 141.0 | ne | - | ‑ S9 ne | S?S? | |
| 55. Koshiki No.2** Japan | 6 | 209.2 | 4 | 104.0 | ‑ S9‑ S10 | S?S? | |
| 56. Koshiki No.3** Japan | ne | - | 3 | 261.3 | ne ‑ S10 | S5S? | |
| 57. Koshiki No.4** Japan | 3 | 99.7 | ne | - | ‑ S9 ne | S?S? | |
| 58. Koshiki No.6** Japan | 6 | 154.2 | 6 | 362.3 | ‑ S9‑ S10 | S5S? | |
| 59. Koshiki No.9** Japan | ne | - | 3 | 143.0 | ne ‑ S10 | S5S? | |
| 60. Mato Anyu** Taiwan | 3 | 488.7 | 3 | 185.0 | ‑ S9‑ S10 | S?S? | |
| 61. Mato Buntan** Taiwan | 6 | 338.8 | 6 | 181.7 | ‑ S9‑ S10 | S3S11 | |
| 62. Mato Peiyu** Taiwan | 3 | 288.3 | 3 | 94.0 | ‑ S9‑ S10 | S1S? | |
| 63. Nagashima No.1** Japan | 3 | 108.0 | 3 | 190.0 | ‑ S9‑ S10 | S?S? | |
| 64. Nagashima No.5** Japan | 3 | 298.3 | 3 | 105.0 | ‑ S9‑ S10 | S?S? | |
| 65. Nagashima No.9** Japan | 3 | 278.7 | ne | - | ‑ S9 ne | S5S? | |
| 66. Nameless** Japan | 3 | 180.3 | 3 | 178.7 | ‑ S9‑ S10 | S?S? | |
| 67. Nejime Buntan** Japan | 4 | 62.5 | ne | - | ‑ S9 ne | S1S? | |
| 68. Omuta No.1** Japan | 6 | 263.3 | 6 | 327.5 | ‑ S9‑ S10 | S1S? | |
| 69. Sekitoyu** Taiwan | 6 | 171.7 | 6 | 176.7 | ‑ S9‑ S10 | S2S? | |
| 70. Shatienyu (NIFTS)** China | 2 | 149.0 | 3 | 212.0 | ‑ S9‑ S10 | S?S? | |
| 71. Shatienyu ZS(KUPFTES)**China | 6 | 147.2 | 5 | 175.6 | ‑ S9‑ S10 | S?S? | |
| 72. Soyu** Taiwan | ne | - | 2 | 149.0 | ne ‑ S10 | S1S2 | |
| 73. Suanyu seedling** China | 3 | 197.7 | ne | - | ‑ S9 ne | S?S? | |
| 74. Taiwan Buntan** Taiwan | 3 | 176.3 | 6 | 105.5 | ‑ S9‑ S10 | S5S11 | |
| 75. Tanegashima No.4** Japan | 3 | 267.0 | 3 | 181.3 | ‑ S9‑ S10 | S5S? | |
| 76. Tanegashima No.9** Japan | 3 | 88.0 | 3 | 166.3 | ‑ S9‑ S10 | S?S? | |
| 77. Tanegashima No.11** Japan | ne | - | 3 | 45.0 | ne ‑ S10 | S?S? | |
| 78. Tanegashima No.12** Japan | 3 | 175.0 | ne | - | ‑ S9 ne | S?S? | |
| 79. Tanegashima No.13** Japan | 4 | 146.8 | 6 | 104.5 | ‑ S9‑ S10 | S?S? | |
| 80. Tanegashima No.15** Japan | ne | - | 3 | 94.7 | ne ‑ S10 | S?S? | |
| 81. Tanegashima No.16** Japan | ne | - | 3 | 179.0 | ne ‑ S10 | S?S? | |
| 82. Tanegashima No.17** Japan | ne | - | 3 | 148.3 | ne ‑ S10 | S?S? | |
| 83. Vietnam No.2** Japan | 3 | 320.0 | 2 | 142.5 | ‑ S9‑ S10 | S?S? | |
| 84. Yatsushiro No.1** Japan | 2 | 157.5 | ne | - | ‑ S9 ne | S1S? | |
| 85. Yatsushiro No.4** Japan | 3 | 287.3 | 6 | 1.5 | ‑ S9 +S10 | S10S? | |
| 86. Yatsushiro No.8** Japan | 3 | 189.0 | 3 | 166.7 | ‑ S9‑ S10 | S?S? | |
| C. maxima complex | |||||||
| 87. Bangkok Buntan (NIFTS)** | ne | - | 3 | 172.3 | ne ‑ S10 | S?S? | |
| 88. Higo Pummelo** | 3 | 244.3 | 2 | 150.0 | ‑ S9‑ S10 | S4S11 | |
| 89. Hino Buntan** | 3 | 127.0 | 3 | 178.7 | ‑ S9‑ S10 | S1S? | |
| 90. Itoshima-bankan** | 3 | 213.0 | 3 | 136.3 | ‑ S9‑ S10 | S5S? | |
| 91. Kessaku** | 3 | 113.0 | 2 | 141.7 | ‑ S9‑ S10 | S?S? | |
| 92. Kochihakuyu** | 3 | 286.3 | 3 | 127.1 | ‑ S9‑ S10 | S?S? | |
| 93. Natsudai* | 6 | 150.2 | ne | - | ‑ S9 ne | SfS2 | |
| 94. Kugatsukan** | 3 | 248.0 | 3 | 149.7 | ‑ S9‑ S10 | S?S? | |
| 95. Morrison* | 3 | 228.7 | 3 | 92.7 | ‑ S9‑ S10 | SfS? | |
| 96. Tanikawa Buntan** | 5 | 213.2 | 3 | 182.0 | ‑ S9‑ S10 | S?S? | |
| C. truncata hort. ex Tanaka (57) | |||||||
| 97. Kaikokan* | ne | - | 3 | 275.7 | ne ‑ S10 | SfS4 | |
| C. pseudogulgul hort. ex Shirai (59) | |||||||
| 98. Shishiyuzu** | 3 | 245.7 | 3 | 266.7 | ‑ S9‑ S10 | S1S? | |
| C. paradisi hort. Macf. (62) | |||||||
| 99. Foster Pink* | 3 | 133.0 | 3 | 183.0 | ‑ S9‑ S10 | SfS? | |
| 100. Mash* | 3 | 114.0 | 3 | 253.3 | ‑ S9‑ S10 | SfS? | |
| 101. Triumph* | 3 | 300.0 | 3 | 162.7 | ‑ S9‑ S10 | SfS? | |
| C. hassaku hort. Tanaka (74) | |||||||
| 102. Hassaku** | 3 | 154.3 | 3 | 118.0 | ‑ S9‑ S10 | S4S5 | |
| C. iwaikan hort. ex Y. Tanaka (75) | |||||||
| 103. Iwaikan** | 3 | 161.5 | 3 | 211.0 | ‑ S9‑ S10 | S?S? | |
| C. tengu hort. ex Tanaka (76) | |||||||
| 104. Tengu* | 3 | 96.0 | 3 | 112.3 | ‑ S9‑ S10 | SfS4 | |
| C. medioglobosa hort. ex Tanaka (77) | |||||||
| 105. Naruto*? | 3 | 183.7 | 3 | 162.3 | ‑ S9‑ S10 | SfS? | |
| C. natsudaidai Hayata (78) | |||||||
| 106. Kawano Natsudaidai* | 3 | 162.0 | ne | - | ‑ S9 ne | SfS2 | |
| C. obovoidea hort. ex I. Tanaka (79) | |||||||
| 107. Kinkoji** | 2 | 71.0 | ne | - | ‑ S9 ne | S4S? | |
| C. otachibana hort. ex Y. Tanaka (80) | |||||||
| 108. Tosa Buntan** | 3 | 177.0 | 3 | 213.3 | ‑ S9‑ S10 | S1S3 | |
| C. ampullaceal hort. ex Tanaka (81) | |||||||
| 109. Hyokan** | 6 | 138.1 | 2 | 135.5 | ‑ S9‑ S10 | S?S? | |
| C. yuge-hyokan hort. ex Tanaka (82) | |||||||
| 110. Yuge-hyokan** | 6 | 135.0 | 3 | 58.3 | ‑ S9‑ S10 | S4S? | |
| C. sulcata hort. ex Takahashi (84) | |||||||
| 111. Sanbokan* | 5 | 206.2 | 4 | 133.7 | ‑ S9‑ S10 | SfS? | |
| C. taiwanica hort. Tanaka et Shimada (87) | |||||||
| 112. Nanshodaidai*? | 1 | 91.0 | ne | - | ‑ S9 ne | SfS? | |
| C. papillaris hort. Blanco (87) | |||||||
| 113. Chizon*? | 3 | 295.3 | ne | - | ‑ S9 ne | SfS? | |
| C. anseikan hort. ex Tanaka | |||||||
| 114. Anseikan** | 3 | 213.0 | 2 | 49.0 | ‑ S9‑ S10 | S1S? | |
| Sour orange and relatives | |||||||
| C. aurantium Linn. (93) | |||||||
| 115. Bonotsu-daidai* | 3 | 329.0 | 6 | 184.5 | ‑ S9‑ S10 | SfS? | |
| 116. Bouquet des Fleurs* | 3 | 330.0 | 3 | 110.0 | ‑ S9‑ S10 | SfS? | |
| 117. Kabusu* | 5 | 0.0 | 6 | 110.5 | +S9‑ S10 | SfS9 | |
| 118. Zadaidai* | 6 | 41.3 | 9 | 123.4 | ‑ S9‑ S10 | SfS11 | |
| C. aurantium × C. paradisi? | |||||||
| 119. Smooth Seville*? | 3 | 330.0 | 3 | 284.7 | ‑ S9‑ S10 | SfS? | |
| C. canaliculata hort. ex Y. Tanaka (99) | |||||||
| 120. Kiku-daidai* | 3 | 0.0 | 3 | 106.0 | +S9‑ S10 | S9S11 | |
| Sweet orange and relatives | |||||||
| C. sinensis Osbeck (100) | |||||||
| 121. Otao orange* | 2 | 132.5 | 3 | 257.7 | ‑ S9‑ S10 | SfS3 | |
| 122. Person Brown* | 6 | 125.0 | 2 | 197.5 | ‑ S9‑ S10 | SfS3 | |
| 123. Ruby Blood* | 3 | 291.7 | 3 | 146.0 | ‑ S9‑ S10 | SfS3sm | |
| 124. Shamouti* | 6 | 233.0 | 2 | 113.0 | ‑ S9‑ S10 | SfS3 | |
| 125. Trovita* | 3 | 139.3 | 3 | 66.7 | ‑ S9‑ S10 | SfS3 | |
| 126. Yoshida Navel* | 3 | 240.3 | 3 | 132.3 | ‑ S9‑ S10 | SfS3 | |
| Yuzu and relatives | |||||||
| C. tamurana hort. ex Tanaka (107) | |||||||
| 127. Hyuganatsu** | 3 | 155.7 | 3 | 148.0 | ‑ S9‑ S10 | S1S? | |
| 128. Orange Hyuga** | 3 | 175.0 | ne | - | ‑ S9 ne | S1S? | |
| C. luteo-turgida Tanaka (109) | |||||||
| 129. Dada** | ne | - | 2 | 93.5 | ne ‑ S10 | S?S? | |
| C. sudachi hort. ex Shirai (115) | |||||||
| 130. Sudachi*? | 3 | 72.0 | 3 | 141.7 | ‑ S9‑ S10 | SfS? | |
| C. pseudoaurantium hort. ex Y. Tanaka (119) | |||||||
| 131. Henka-mikan*? | ne | - | 3 | 115.0 | ne ‑ S10 | SfS? | |
| C. sphaerocarpa hort. ex Tanaka (121) | |||||||
| 132. Kabosu* | 3 | 294.7 | 3 | 95.0 | ‑ S9‑ S10 | SfS4 | |
| Mandarin and relatives | |||||||
| C. nobilis cv. King × C. deliciosa | |||||||
| 133. Encore* | ne | - | 3 | 193.3 | ne ‑ S10 | SfS? | |
| C. nobilis complex | |||||||
| 134. Oshima-mikan Hakunikukei*? | ne | - | 2 | 79.5 | ne ‑ S10 | SfS? | |
| 135. Kusakunebu*? | 3 | 84.0 | 3 | 158.7 | ‑ S9‑ S10 | SfS? | |
| C. unshiu (L1)* + C. obovoidea (L2, L3)** | |||||||
| 136. Kinkoji Unshu* | 3 | 93.3 | ne | - | ‑ S9 ne | SfS4+S4S? | |
| C. genshokan hort. ex Tanaka (132) | |||||||
| 137. Genshokan* | 3 | 167.3 | 3 | 152.3 | ‑ S9‑ S10 | SfS? | |
| C. tangerina hort. ex Tanaka (133) | |||||||
| 138. Dancy* | 3 | 110.3 | 3 | 147.3 | ‑ S9‑ S10 | SfS? | |
| C. tangerina × C. paradisi | |||||||
| 139. Seminole* | 3 | 232.7 | 3 | 317.3 | ‑ S9‑ S10 | SfS? | |
| C. clementina hort. ex Tanaka (134) | |||||||
| 140. Clementine** | 2 | 85.0 | ne | - | ‑ S9 ne | S3S11 | |
| C. tachibana Tanaka (143) | |||||||
| 141. Tachibana No.1 (NIFTS)*? | 3 | 66.7 | 3 | 82.7 | ‑ S9‑ S10 | SfS? | |
| 142. Tachibana No.2 (KUPFTES)*? | 3 | 38.0 | 3 | 221.0 | ‑ S9‑ S10 | SfS? | |
| C. depressa Hayata (153) | |||||||
| 143. Hijakunibu*? | ne | - | 3 | 116.7 | ne ‑ S10 | SfS? | |
| 144. Kaachi*? | 2 | 91.8 | 2 | 140.0 | ‑ S9‑ S10 | SfS? | |
| 145. Ogimi Kugani*? | ne | - | 2 | 100.5 | ne ‑ S10 | SfS? | |
| C. flaviculpus hort. ex Tanaka | |||||||
| 146. Ogonkan (Kimikan)** | 2 | 105.5 | 2 | 223.0 | ‑ S9‑ S10 | S4S? | |
zSelf-incompatibility (**) and semi-self-incompatibility (*) in most accessions are based on the report of Ngo et al. (2019), and the others depend on the following reports: present study, Kim et al. (2010, 2011, 2020), Soost and Roose (1969), Yamamoto et al. (2006), Vardi et al. (2000) and Zhou et al. (2018).
yFresh S9 pollen was collected from HBS3, HBS4, HBS19 and HBS25 with the S9S9 genotype, while S10 pollen was collected from HBS2, HBS11 and HBS20 with the S10S10 genotype. All the pollinated pistils were collected 8 days after pollination. Data for accession Nos. 29, 33-40, 98, 102, 106, 108, 110, 114, 117, 127 and 143 include those reported by Kim et al. (2010).
xDetermined S genotypes except for those determined in the present study is based on the report of Kim et al. (2010) for S9 and S10, Kim et al. (2011) for S1 and S2, Zhou et al. (2018) for S4 and S5, and Kim et al. (2020) for S3 and S11.
Determination of Citrus Accessions with S10 Pollen
When the S10 pollen of ‘Hirado Buntan’ pummelo homozygous S1 seedlings (S9S9) was pollinated to 127 Citrus accessions, many S9 pollen tubes penetrated into the lower styles of 122 accessions (Table 1 and Fig. 1). The average number of pollen tubes observed in the lower styles of the 122 accessions ranged from 45.0 to 382.7. This result suggested that these accessions show cross compatibility with S10 pollen and do not have an S10 allele. In five of the 127 accessions, no or only a few pollen tubes were observed in the lower styles: 0.0 for Amakusa No. 6 pummelo (Citrus maxima), 9.0 for the ‘Hirado Buntan’ pummelo, 0.0 for the ‘Bangkok Buntan’ (KPFTES) pummelo (Citrus maxima), 1.7 for the Kawanabe No. 2 pummelo (Citrus maxima), and 1.5 for the Yatsushiro No. 4 pummelo (Citrus maxima). This result suggested that these pummelo accessions show cross incompatibility with S10 pollen and have an S10 allele. Except for accessions belonging to the pummelo group, those that have an S10 allele were not detected in the other groups of Citrus accessions examined.
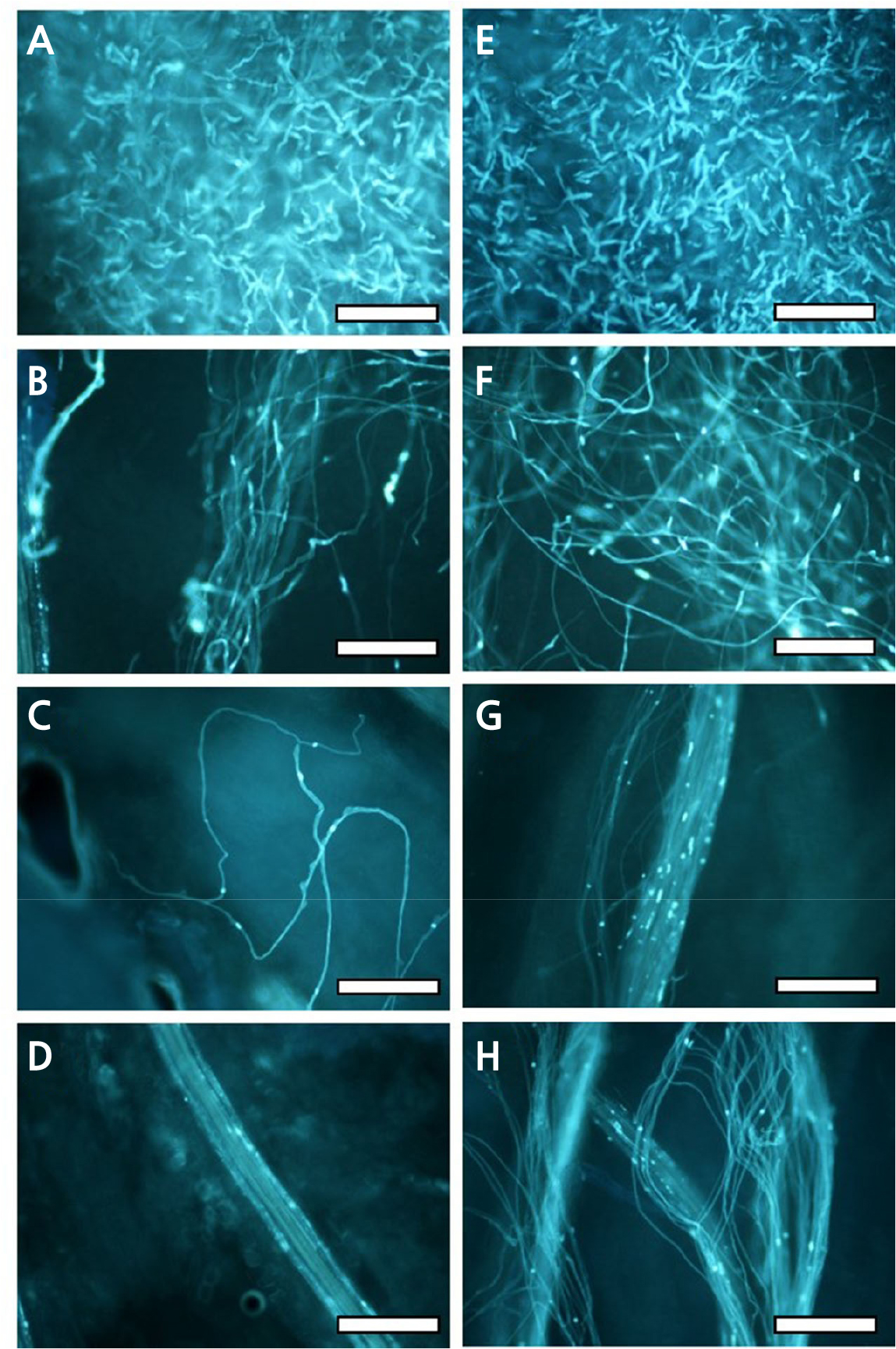
Fig. 1.
Pollen tube arrest in the pistil of ‘Hirado Buntan’ (S9S10) pollinated with HBS2 (S10S10) (A-D), and normal pollen tube growth in the pistil of ‘Hassaku’ (S4S5) pollinated with HBS3 (S9S9) (E-H). A and E: Stigma; B and F: upper part of style; C and G: middle part of style; D and H: lower part of style. Note the multiple pollen tubes in each section of the style from the stigma to the style base in right photos and the reduction of the number of pollen tubes from multiple (A) to zero (D) in the left photos. In photo D, no pollen tube is seen; the pollen tube-like structure emitting strong fluorescence is the phloem of conductive tissue in the style base. Bars indicate 0.1 mm.
Variation of the Number of Pollen Tubes in Lower Styles of Back-cross-incompatible Pistils (S9S10) Pollinated with ‘Hirado Buntan’ (S9S10)
In the upper styles of back-crossed pistils (S9S10) with ‘Hirado Buntan’ (S9S10), more than 200 pollen tubes per style were observed, while in the lower styles the average number of pollen tubes ranged from 2.5 for HBS27 to 8.0 for HBS5 with the average number of 5.8 per style (Table 2). The average number of 5.8 pollen tubes per style was almost the same as that in self-pollination of ‘Hirado Buntan’ reported by Ngo et al. (2019) (Table 2).
Table 2.
Small variation for SI in lower styles between self- and back-crosses pollinated with ‘Hirado Buntan’ pummelo
|
Pistillate parent × pollen parent (S genotype) | Pollen genotype | No. of pistils examined | No. of pollen tubes in indicated part of style | |
| Upper | Lower | |||
| Hirado Buntan (S9S10) × Hirado Buntan (S9S10)z | S9 and S10 | 3 | >150.0 | 5.7 |
| HBS5 (S9S10) × Hirado Buntan (S9S10) | S9 and S10 | 2 | >300.0 | 8.0 |
| HBS15 (S9S10) × Hirado Buntan (S9S10) | S9 and S10 | 2 | >300.0 | 7.0 |
| HBS18 (S9S10) × Hirado Buntan (S9S10)y | S9 and S10 | 3 | >200.0 | 7.3 |
| HBS25 (S9S10) × Hirado Buntan (S9S10) | S9 and S10 | 2 | >300.0 | 4.0 |
| HBS27 (S9S10) × Hirado Buntan (S9S10) | S9 and S10 | 2 | >300.0 | 2.5 |
| Average number of pollen tubes in lower styles | 5.8 | |||
zCited from Ngo et al., 2019.
yCited from Kim et al., 2010.
Variation of the Number of Pollen Tubes in the Lower Styles of Incompatible Pistils of Citrus Plants Carrying S9 and/or S10 Alleles When Pollinated with S9 or S10 Pollen
In the upper styles of all cross-incompatible pistils pollinated with S9 pollen, more than 72 pollen tubes per style were observed (Table 3). In the lower styles of all pistils pollinated with S9 pollen, the average number of pollen tubes observed ranged from 0 for Kabusu (SfS9) × HBS3 (S9S9) to 8.5 for HBS4 (S9S9) × HBS3 (S9S9) with the average of 2.3 per style (Table 3). In the lower styles of all pistils pollinated with S9 pollen from HBS3 (S9S9), the average number of pollen tubes was 8.5, 0.7 and 5.0 per lower style of HBS4, HBS7 and HBS18, respectively, and 0.8, 0.7 and 0.6 for ‘Hirado Buntan’, ‘Kabusu’ and Amakusa No.6 pummelos (Table 3). HBS19 (S9S9) × HBS25 (S9S9) and Hirado Buntan (S9S10) × HBS4 (S9S9) showed 1.0 and 2.0 pollen tubes per lower style.
Table 3.
Degree of pollen tube arrest for S9 and S10 pollens
|
Pistillate parent × pollen parent (S genotype) | Pollen genotype | No. of pistils examined | No. of pollen tubes in indicated part of style | |
| Upper | Lower | |||
| HBS4 (S9S9) × HBS3 (S9S9)z | S9 | 2 | >150.0 | 8.5 |
| HBS7 (S9S9) × HBS3 (S9S9)z | S9 | 3 | >150.0 | 0.7 |
| HBS18 (S9S10) × HBS3 (S9S9)z | S9 | 2 | >150.0 | 5.0 |
| HBS19 (S9S9) × HBS25 (S9S9) | S9 | 3 | >150.0 | 1.0 |
| Hirado Buntan (S9S10) × HBS3 (S9S9) | S9 | 7 | >200.0 | 0.8 |
| Hirado Buntan (S9S10) × HBS4 (S9S9) | S9 | 1 | >200.0 | 2.0 |
| Kabusu (SfS9) × HBS3 (S9S9) | S9 | 5 | 72.0 | 0.0 |
| Amakusa No.6 (S9S10) × HBS3 (S9S9) | S9 | 3 | 249.3 | 0.3 |
| Average number of S9 pollen tubes in lower styles | 2.3 | |||
| Amakusa No.6 (S9S10) × HBS2 (S10S10) | S10 | 3 | 180.7 | 0.0 |
| Yatsushiro No.4 (S10S11) × HBS2 (S10S10) | S10 | 3 | >200.0 | 0.0 |
| Bangkok Buntan (S10S?) × HBS2 (S10S10) | S10 | 3 | >200.0 | 0.0 |
| Kawanabe No.2 (S5S10) × HBS2 (S10S10) | S10 | 3 | >200.0 | 1.7 |
| Hirado Buntan (S9S10) × HBS2 (S10S10) | S10 | 4 | >200.0 | 9.0 |
| Average number of S10 pollen tubes in lower styles | 2.1 | |||
zCited from Kim et al., 2010.
In the upper styles of all cross-incompatible pistils pollinated with S10 pollen from HBS2 (S10S10), more than 180.7 pollen tubes per style were observed (Table 3). In the lower styles of pistils pollinated with S10 pollen, the average number of pollen tubes observed was the highest (9.0) in ‘Hirado Buntan’, followed by 1.7 in Kawanabe No. 2 (S5S10) and 0 in Amakusa No.6 (S9S10), Yatsushiro No.4 (S10S11) and ‘Bangkok Buntan’ (S10S?) pummelos with the average number of 2.1 pollen tubes per lower style (Table 3). The average number of 2.1 per lower style was almost the same as that of 2.3 in cross-incompatible pistils pollinated with S9 pollen.
Discussion
Distribution of Citrus Accessions with S9 and/or S10 Alleles
In this study, S gene alleles were found in accessions belonging to pummelo and a pummelo relative for the S9 allele and those belonging to pummelos for the S10 allele. This suggests that S9 and S10 alleles have their origin in pummelo and not in mandarin. In the previous study with S9 and S10 pollens, neither the S9 nor the S10 allele was found in seven Hirado Island pummelo accessions estimated to be relatives of the ‘Hirado Buntan’ pummelo (Kim et al., 2011). This suggests that the S9 and S10alleles of ‘Hirado Buntan’ are unique to the Hirado Island pummelo group. In the present study, we found that S9 and S10alleles exist in the Amakusa No. 6 pummelo, which was collected from an old tree located at Yunuki in the Amakusa Islands, Kumamoto Prefecture. The Amakusa No. 6 pummelo has a morphology similar to that of ‘Hirado Buntan’ in the tree, flowers, and fruit. The old tree was seeded and likely was not grafted, and the self-incompatibility reaction in the pistils to S10 pollen differed from that observed in ‘Hirado Buntan,’ i.e., 0.0 for the Amakusa No. 6 pummelo and 9.0 for ‘Hirado Buntan’ (Table 1). It may be said that the Amakusa No. 6 pummelo and ‘Hirado Buntan’ are not the same plant but different genotypes.
‘Kabusu,’ one of four sour orange accessions examined, has an S9allele (Table 1) as suggested by Kim et al. (2011), while three of the four accessions have no S9allele. Kim et al. (2020) reported that the ‘Zadaidai’ sour orange, one of the four sour orange accessions examined in this study, has an S11 allele. On the other hand, two of the four sour orange accessions examined have neither an S9allele nor an S10allele. Although it has been reported that the sour orange originated from an F1 hybrid between a pure pummelo and mandarin (Wu et al., 2014) or is an offspring of a cross between lemon and the F1 of a pummelo and mandarin cross (Shimizu et al., 2016), it has also been reported that sour orange accessions show various differences not only in morphology but also in molecular markers (Siragusa et al., 2006). Thus, the S allele difference among ‘Kabusu’ (SfS9), ‘Zadaidai’ (SfS11), and two of the four sour orange accessions (‘Bonotsu-daidai’ and ‘Bouquet des Fleurs’) may suggest that they are genetically different cultivars, not clones in which S allele mutation occurred during the long history of cultivation. To verify the genotype of the original or pure sour orange, further assessment of common or standard sour oranges is necessary. In the present study, we demonstrated that ‘Kikudaidai,’ a very close relative of sour orange, has an S9 allele, while Kim et al. (2020) reported that ‘Kikudaidai’ has an S11 allele. This suggests the possibility that ‘Kikudaidai’ (S9S11) was produced by direct crossing between ‘Kabusu’ and ‘Zadaidai’ or by crossing between a plant with ‘Kabusu’ and one with ‘Zadaidai’ in their pedigrees.
In addition to Amakusa No. 6 (S9S10) and ‘Hirado Buntan’ (S9S10), ‘Bangkok Buntan’ (KPFTES), the Kawanabe No. 2 pummelo, and Yatsushiro No. 4 pummelo have an S10allele. Although the S10allele is distributed in pummelo accessions at a higher rate than the S9allele, it does not appear in the Citrus accessions except for pummelo and ‘Kikudaidai’. These results suggest that the distribution of Citrus accessions with S9 and/or S10alleles is limited to pummelo and sour orange, probably because of the low fruit quality of these accessions, resulting in limited expansion of Citrus accessions with S9 and/or S10alleles during the long history of cultivation of pummelo and sour orange—about 500 years (estimation by authors) and about 2000 years in Japan (Tanaka, 1948b) respectively, and for several thousand years in other parts of the world. A ‘Hirado Buntan’ pummelo with high fruit quality was introduced in Japan in 1846 (Tanaka, 1948a), and thus, the introgression of S9 and/or S10alleles into the Citrus accessions is thought to be very limited in Japan.
Characteristics of the S Allele Frequency for S9 and S10 Alleles
The rates of Citrus accessions with Sn alleles except for Sf alleles have been reported for the S1 and S2 (Kim et al., 2011), S3 (Kim et al., 2020), S4 and S5 (Zhou et al., 2018), and S11 (Kim et al., 2020) alleles of pummelo origin. According to these reports, the rates of Citrus accessions with Sn alleles except for the Sf allele are 29.9% for S1, 21.3% for S2, 17.7% for S3, 9.4% for S4, 6.3% for S5, and 13.2% for S11. The present result for the rates of Citrus accessions with S9 and S10 alleles are 3.1% (4 of 132 accessions) and 3.9% (5 of 129 accessions), respectively. When six sweet orange accessions and two Hyuganatsu accessions (Table 1) are counted as one genotype each, the rates of Citrus genotypes with S9 and S10 alleles are 3.2% (4 of 126 accessions) and 3.9% (5 of 127 accessions), respectively. The pummelo accessions had S9 and S10 alleles at higher rates of 2.9% (2 of 70 accessions examined) and 7.0% (5 of 71 accessions examined), respectively. The rates for S9 and S10 alleles are the lowest and second lowest among the eight rates determined. The rates of pummelo accessions with Sn alleles except for the Sf allele are 34.4% for S1, 26.4% for S2, 12.2% for S3, 1.4% for S4, 11.8% for S5, and 33.3% for S11. These S allele frequencies are 17.2% for S1, 13.2% for S2, 6.1% for S3, 0.7% for S4, 5.9% for S5, and 16.7% for S11. The rates of pummelo accessions with S9 and S10 alleles are 2.9% and 7.0%, respectively. Therefore, the S9 and S10 allele frequencies are 1.8% and 2.3%, respectively. The S9 and S10 allele frequencies are second and third lowest among the eight S allele frequencies in pummelo. The total of the eight S allele frequencies in pummelo is 64.6%, from which it is roughly estimated that more than twelve S alleles are estimated to exist in pummelo.
Differences in the Self-incompatibility Reaction in Lower Styles between S9 and S10 Alleles
Ngo et al. (2019) classified the self-incompatibility reaction into several types by the degree of pollen tube arrest in the upper, middle, and lower parts of the styles. They classified self-incompatibility in ‘Hirado Buntan’ into an MMH type (Moderate in stigma, Moderate in upper style, High in middle style). It exhibited, however, a relatively slow or mild self-incompatibility reaction in the lower styles with an average penetration of 5.7 pollen tubes per style and in ovaries with an average penetration of 3.7 pollen tubes per ovary, although the self-pollinated fruit is seedless (Ngo et al., 2019). Yamamoto et al. (2006) observed a similar phenomenon in the self-pollination of ‘Hirado Buntan’ carrying S9 and S10 pollen. The present data for back-cross-incompatibility in pollinations with ‘Hirado Buntan’ pummelo showed a slow self-incompatibility reaction in the lower part of styles with an average penetration of 5.8 pollen tubes per lower style, similar to the value of self-pollination of ‘Hirado Buntan’ (Table 2). The result suggests that the same extent of mild SI reaction in the lower part of styles occurs in all five back-crosses. This also suggests the possibility that the mild SI reaction in the lower part of styles of ‘Hirado Buntan’ is an inherited trait.
On the other hand, we postulated that the extent of mild SI reaction in the lower part of styles will be different between pollinations with S9 pollen and S10 pollen, since when ‘Hirado Buntan’ (S9S10) was pollinated with S9 pollen of ‘Hirado Buntan’ S1 seedlings (S9S9), 1.0 S9 pollen tube on average per lower style was observed, whereas when pollinated with S10 pollen of another ‘Hirado Buntan’ S1 seedling (S10S10), 9.0 S10 pollen tubes on average per lower style were observed (Table 1). The average number of S9 pollen tubes of HBS3 and HBS4 penetrating into the lower style of ‘Hirado Buntan’ was 1.1 and 2.0, respectively, while the average number of S10 pollen tubes of HBS2 penetrating into the lower style of ‘Hirado Buntan’ was 9.0 (Table 3). The number of S9 and S10 pollen tubes may suggest that in the lower style of ‘Hirado Buntan’, pollen tubes carrying the S10 allele show a slower incompatibility reaction than those carrying the S9 allele. Hiratsuka et al. (2012) reported that in the Japanese pear [Pyrus pyrifolia (Burm.f.) Nakai], the degree of self-incompatibility is different between the pollen grains carrying different S alleles. In all crosses with S9 or S10 pollen, however, no difference was detected in the number of pollen tubes in the lower styles between S9 and S10 pollen with the average of 1.4 S9 pollen tubes per lower style and 2.2 S10 pollen tubes per lower style (Table 3). The slow or mild self-incompatibility reaction in the lower styles of self-pollinated ‘Hirado Buntan’ and the back-crossed S1 seems to be specific to the pollination with ‘Hirado Buntan’. On the other hand, it may be said generally that there is no clear difference in the mild self-incompatibility reaction in the lower styles between S9 and S10 pollens; the extent of the mild self-incompatibility reaction in the lower styles will be different in pollinations with S9 or S10 pollen.
Conclusion
Of the 146 Citrus accessions including 82 pummelo accessions examined, self-incompatibility genotypes have been completely ascertained or determined in ‘Kabusu’ sour orange (SfS9), a ‘Kikudaidai’ sour orange relative (S9S11) and two local pummelo plants. The S allele frequencies for S9 and S10 alleles of pummelo origin are very low in pummelo accessions and eventually in Citrus accessions compared to those of the other S alleles of pummelo origin reported, suggesting pummelo plants with S9 and S10 alleles did not contribute to the evolution and development of pummelos and eventually Citrus cultivars, except for ‘Hirado Buntan’ pummelo and sour oranges that are used in the commercial production of fresh fruit and root stocks. On the other hand, the S9 allele may be a good marker to detect cultivars and hybrids with ‘Hirado Buntan’ and sour oranges in their pedigrees. The mild self-incompatibility reaction seen in the lower style of self-pollinated ‘Hirado Buntan’ was also confirmed in the lower styles of the S1 plants pollinated with ‘Hirado Buntan’. However, there is no clear difference in the mild self-incompatibility reaction in the lower styles between S9 and S10 pollen. Finally, we examined the degree of self-incompatibility between S9and S10alleles in the lower part of styles of S1 seedlings of ‘Hirado Buntan’ and Citrus accessions with S9and/or S10 alleles. The result indicated no difference in the reaction between the two alleles.


