Introduction
Materials and Methods
Sample Source and Total RNA Preparation
Reverse-Transcription and Polymerization Chain Reaction with NeLV-Specific Primers
Nucleotide Sequencing and Phylogeny Estimation
Results
Discussion
Introduction
Ornamental flower bulbs are important floriculture crops. Hippeastrum, commonly known as amaryllis, is a genus consisting of 90 species and over 600 hybrids and cultivars of perennial herbaceous bulbs in the Amaryllidaceae family. In Korea, amaryllis, an ornamental flowering plant, has been imported from South Africa, Indonesia, and China (2019 Year Book of Plant Quarantine Statistics, Animal and Plant Quarantine Agency, 2020). Some viruses have been reported to infect amaryllis plants when they are tested using leaves. Commonly detected viruses in Hippeastrum species are nerine latent carlavirus (NeLV), narcissus yellow streak potyvirus (NYSV), and hippeastrum mosaic potyvirus (HiMV) (Wylie and Jones, 2012). In addition, cucumber mosaic virus (CMV) has been detected in amaryllis plants in Iraq (Adhab and Al-Ani, 2011). NeLV has recently been found in amaryllis bulbs during quarantine inspection in Korea was reported in a publication from the quarantine agency (2019 Year Book of Plant Quarantine Statistics, Animal and Plant Quarantine Agency, 2020). NeLV is in the genus Carlavirus within the family Betaflexiviridae according to the International Committee on Taxonomy of Viruses. Generally, virus-infected bulbs should be eliminated during quarantine. Since NeLV is considered a provisionally regulated virus in the quarantine process in Korea, amaryllis bulbs should be diagnosed before being imported and planted. These bulbs are imported from abroad and propagated vegetatively, and the initial virus detection should be performed using a bulb at the dormant stage. Because the viruses survive in the dormant bulb, they may be transported across long distances via international trade (Pearson et al., 2009). The vegetative means of propagating bulbs favor the accumulation and dissemination of viruses (Wylie and Jones, 2012; Kim et al., 2019; Lee and Lee, 2019). Therefore, amaryllis bulb tissues were used to investigate the contagion of NeLV in this study. NeLV was first characterized in Nerine bowdenii (Maat et al., 1978), and it has since been reported in Hippeastrum, Narcissus, Lycoris, and other Nerine species in the family Amaryllidaceae (Pham et al., 2010; Wylie and Jones, 2012). NeLV has been detected in the blood lily plant (Haemanthus multiflorus) by serological and molecular identification in Taiwan (Chen et al., 2011), and it has also been identified in Haemanthus albiflos in the UK (Monger and Eden, 2019). NeLV has also been reported in the United States of America (Jordan et al., 2018). However, NeLV has not yet been reported in any bulb ornamental plants in Korea, except for in quarantine reports. Therefore, we attempted to determine NeLV infection using amaryllis bulbs purchased from a commercial market. In this study, amaryllis plants were investigated using reverse-transcription polymerase chain reaction (RT-PCR) to detect NeLV. This study represents the first finding of NeLV in amaryllis bulbs in Korea.
Materials and Methods
Sample Source and Total RNA Preparation
Amaryllis (Hippeastrum hybridum) bulbs were purchased from a commercial market (Yangjae Flower Market, Seoul, Korea). Amaryllis cultivar ‘Red Lion’ (10 bulbs) and ‘Alfresco’ (5 bulbs), approximately 6 cm in diameter, were subjected to RT-PCR detection of NeLV. For total RNA extraction, fresh bulb tissue (150 mg) was frozen in liquid nitrogen and ground to a fine powder. The homogenized bulb tissue was transferred to a microcentrifuge tube, and 200 µL of RNA extraction buffer (50 mM Tris-Cl pH 8.0, 0.1% SDS, 10 mM EDTA pH 8.0, 1% β-mercaptoethanol) and 200 µL of PCI (phenol:chloroform:isoamyl alcohol = 25:24:1) were added. After centrifugation, the supernatant was transferred to a fresh microcentrifuge tube, and an equal volume of 95% ethyl alcohol was added and mixed. After centrifugation, the RNA pellet was washed with 70% ethanol. Finally, the pellet was air-dried and then resolved in 100 µL sterilized distilled water. Quality and quantity were checked using a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA) and stored at −80°C.
Reverse-Transcription and Polymerization Chain Reaction with NeLV-Specific Primers
Complementary DNA (cDNA) was synthesized at 42 °C for 60 min in a reaction mixture (20 µL) containing 5× reaction buffer (250 mM Tris-HCl pH 8.3, 250 mM KCl, 20 mM MgCl2, 50 mM DTT), 1 µL of 2.5 mM dNTP mix (Takara, Japan), 0.5 µL of 400 µg/µL random hexamer (Qiagen, Germany), 4 µL of total RNA (1 µg), and 100 U of RevertAid reverse transcriptase (Thermo Scientific, USA). The primers specific for NeLV detection were designed based on the sequences of the NeLV genome covering 612 bp of the coat protein (CP) gene (891 bp) obtained from the National Center for Biotechnology Information (NCBI) database (Table 1). The specificity of the designed primer sets, including the internal control gene (18S rDNA), was verified using NCBI primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/ primer-blast/). After the RT step, PCR was performed in a reaction mixture (30 µL) containing 10× PCR buffer (100 mM Tris-HCl (pH 8.3 at 25°C, 500 mM KCl, 15 mM MgCl2), 1 µL of 2.5 mM dNTP mix, 1 µL of 10 pmol primer sets, 5 µL of cDNA, and 1 U of Taq polymerase (Takara, Japan). PCR was performed in a thermal cycler (T100™ Thermal Cycler, Bio-Rad, USA) under the following conditions: 3 min at 95°C, followed by 25 cycles at 95°C for 30 s, 60°C for 30 s, 72°C for 60 s, and finally 5 min at 72°C. After the PCR reaction, 1 µL of PCR products was separated on a 2.5% agarose gel at 100 V for 25 min. The gel was stained with ethidium bromide and visualized using a gel doc system (Gel Doc XR+ System, Bio-Rad, USA).
Table 1.
List of primers used for RT-PCR to detect nerine latent virus
| Primer | Sequence (5'-3') |
Length (bp) |
Tm (°C) | Expected size (bp) |
Genome region | Reference |
| NeLV F | CCT ATG GAT GAG ATT AGRz CCG | 21 | 59.4 | 612 | CP partial | JX880024 |
| NeLV R | GCA ACC ATA CAG CCC ACC | 18 | 58.4 | 612 | ||
| 18S F | CTG CGA ATG GCT CAT TAA ATC AGT TAT AG | 29 | 66.2 | 530 | 18S rDNA partial | JQ283909 |
| 18S R | GCA ACA ACT TAA ATA TAC GCT ATT GGA GC | 29 | 66.2 | 530 |
Nucleotide Sequencing and Phylogeny Estimation
PCR products were cloned directly into the pGEM-T Easy vector using T4 ligase (Promega, USA) and transformed into Escherichia coli strain DH5α competent cells (RBC, Taiwan). Selected plasmids were sequenced by standard sequencing at Macrogen Inc. (Seoul, Korea) and analyzed using the BLAST tool at NCBI (http://www.ncbi.nlm.nih.gov/). Consensus sequences were determined by sequencing three independent bulbs for each cultivar three times. The nucleotide sequences of the PCR products were compared with those of NeLV isolates derived from Amaryllidaceae species and other carlaviruses present in the NCBI database. Sequences from diverse NeLV isolates for sequence alignment were trimmed for comparison with the NeLV isolates obtained in this study. The DNA sequences were aligned using ClustalW in MEGA software (version 10.1.8; PA, USA). Pairwise alignment and multiple alignment were performed with a gap-opening penalty of 15, a gap extension penalty of 6.66, a transition weight of 0.5. The display divergence cut-off value was set to 30%. Based on nucleotide sequences, a phylogenetic tree was constructed by the neighbor-joining method using MEGA software. The dataset was subjected to 1,000 bootstrap replicates.
Results
NeLV detection by RT-PCR was performed using NeLV-specific primers from two cultivars, ‘Red Lion’ and ‘Alfresco’. PCR products were amplified with an expected size of 612 bp, and the internal control was amplified as expected (Fig. 1 and Table 1). Fifteen bulb samples subjected to RT-PCR appeared to be amplified by NeLV-specific primers; therefore, PCR fragments were cloned and sequenced, and their nucleotide sequences were compared to those of NeLV isolates in the database. The determined nucleotide sequences were 612 bp in the CP region of the NeLV genome, according to nucleotide BLAST in NCBI. Nucleotide sequence analysis showed no significant variation among the NeLV isolates from those in this study. The NeLV isolates and carlavirus members analyzed in this study are summarized in Table 2. NeLV isolates used for comparison with the nucleotides analyzed in this study comprised species of Amaryllidaceae. The sequences of the ‘Alfresco’ isolate (NeLV-HA) and ‘Red Lion’ (NeLV-HR) shared 98.5% and 96.7% identity, respectively, with the Taiwan isolate derived from the Hippeastrum host (Table 2). NeLV-HA and NeLV-HR shared 97.2% and 97.5% identity, respectively, with the Netherlands isolate derived from Hippeastrum. Two isolates (NeLV-HA and NeLV-HR) in this study showed 97% sequence identity with each other. When Amaryllidaceae species such as Nerine, Narcissus, Crinum, Lycoris,and Haemanthus isolates of NeLV were compared to NeLV-HA and NeLV-HR, the sequence identity was above 93.9% (Table 2). NeLV has been reported to specifically infect species only in Amaryllidaceae. Other virus species in the carlavirus group were used for comparison, and the results revealed that sequence identity in the analyzed partial CP region ranged from 49.7% to 59% (Table 2). This result indicates that there is high homology among NeLV isolate-derived species within the Amaryllidaceae family.
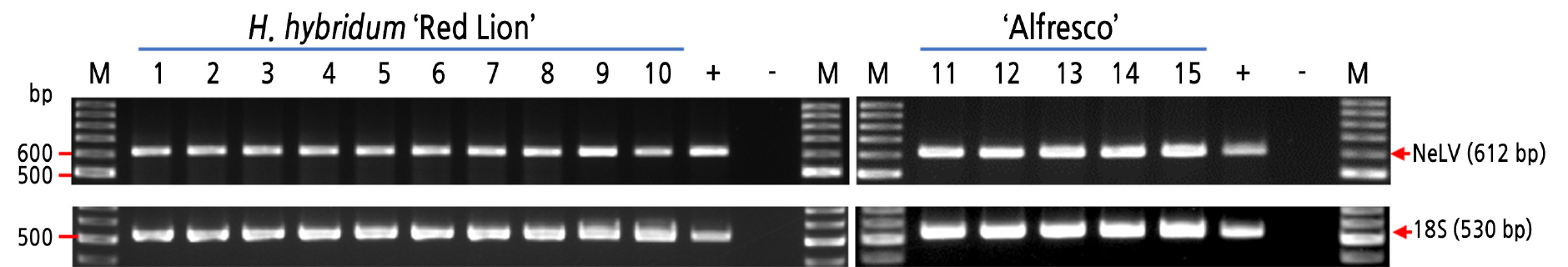
Fig. 1.
RT-PCR analysis using NeLV-specific primers to identify NeLV infection in the bulbs of Hippeastrum hybridum (upper panel); 18S rDNA amplification was used as an internal control (lower panel). M, 100-bp DNA ladder marker; 1-10, H. hybridum ‘Red Lion’; 11-15, H. hybridum ‘Alfresco’; (+), positive control; (‒), sterile distilled water instead of cDNA.
Table 2.
Percent identities of the nucleotide sequences of NeLV corresponding to the partial coat protein gene isolated from H. hybridum ‘Alfresco’ and ‘Red Lion’ with other NeLV isolates, other carlaviruses, and outgroup species
| Virus namez |
GenBank accession no. | Host species | Country isolated | Identity (%) | |
| NeLV-HAy | NeLV-HRx | ||||
| NeLV | MW728097 | H. hybridum ‘Alfresco’ | Korea | 100 | 97.0 |
| NeLV | MW711816 | H. hybridum ‘Red Lion’ | Korea | 97.0 | 100 |
| NeLV | HM119495.1 | Hippeastrum sp. | Netherlands | 97.2 | 97.5 |
| NeLV | HM119498.1 | Nerine sp. | Netherlands | 97.4 | 97.7 |
| NeLV | JQ395044.1 | Narcissus sp. | Australia | 97.4 | 97.7 |
| NeLV | JX524884.1 | Narcissus sp. | Taiwan | 97.5 | 97.9 |
| NeLV | JX880024.1 | Hippeastrum sp. | Taiwan | 98.5 | 96.7 |
| NeLV | JX978442.1 | Lycoris aurea | Taiwan | 96.4 | 96.4 |
| NeLV | KM396460.1 | Haemanthus multiflorus | Taiwan | 95.6 | 95.9 |
| NeLV | MK085064.1 | Haemanthus albiflos | UK | 97.2 | 97.5 |
| NeLV | MG012804.1 | Crinum sp. | USA | 94.3 | 93.9 |
| NeLV | NC_008552 | Narcissus sp. | Germany | 96.4 | 97.4 |
| CVNV | NC_009764.1 | Verbena officinalis | USA | 55.2 | 54.9 |
| GaLV | LC485538.1 | Valeriana fauriei | Japan | 50.5 | 50.2 |
| GCLV | KF010516.1 | Allium sativum | India | 58.7 | 59.2 |
| HipLV | NC_011540.1 | H. hybridum | Taiwan | 50.3 | 50.0 |
| HpLV | EF394784.1 | Humulus lupulus | China | 50.3 | 50.5 |
| KLV | NC_013006.1 | Kalanchoe blossfeldiana | Denmark | 49.7 | 50.8 |
| LSV | AJ831417.2 | Lilium sp. | India | 53.1 | 52.6 |
| NCLV | NC_008266.1 | Narcissus tazetta var. chinensis | China | 51.8 | 50.8 |
| PhlVM | NC_043083.1 | Phlox | USA | 51.5 | 51.0 |
| PVS | KY523844.1 | Solanum tuberosum | Iran | 52.8 | 53.4 |
| RCVMV | NC_012210.1 | Cicer arietinum L. | USA | 53.4 | 53.9 |
| SLV | MF776530.1 | Allium sativum | Korea | 58.4 | 59.0 |
| PVX | NC_011620.1 | Nicotiana tabacum | 48.4 | 48.3 | |
zAbbreviations of virus names in GenBank. CVNV, Coleus vein necrosis virus; GaLV, Gaillardia latent virus; GCLV, Garlic common latent virus; HipLV, Hippeastrum latent virus; HpLV Hop latent virus; KLV, Kalanchoe latent virus; LSV, Lily symptomless virus; NCLV, Narcissus common latent virus; NeLV, Nerine latent virus; PhlVM, Phlox virus M; PVP, Potato virus P; PVS, Potato virus S; PVX, Potato virus X; RCVMV, Red clover vein mosaic virus; SLV, Shallot latent virus.
To determine the genetic relationship among NeLV isolates from Amaryllidaceae species and other carlaviruses, phylogeny was generated from sequence alignment using the sequences from isolates analyzed in this study (Table 2 and Fig. 2). In the phylogenetic tree, NeLV isolates from Hippeastrum, Lycoris, Crinum, Narcissus, and Nerine were grouped in the same cluster as NeLV-HA and HR. Other carlaviruses were distinctively differentiated from NeLV isolates; therefore, virus isolates obtained in this study could be classified as NeLV according to nucleotide sequence and phylogeny analysis. Except for potato virus X (PVX) as a potexvirus outgroup within Alphaflexiviridae, shallot latent virus (SLV), garlic common latent virus (GCLV), and coleus vein necrosis virus (CVNV) were more closely related to NeLV isolates than other carlaviruses based on the neighbor-joining tree (Fig. 2).
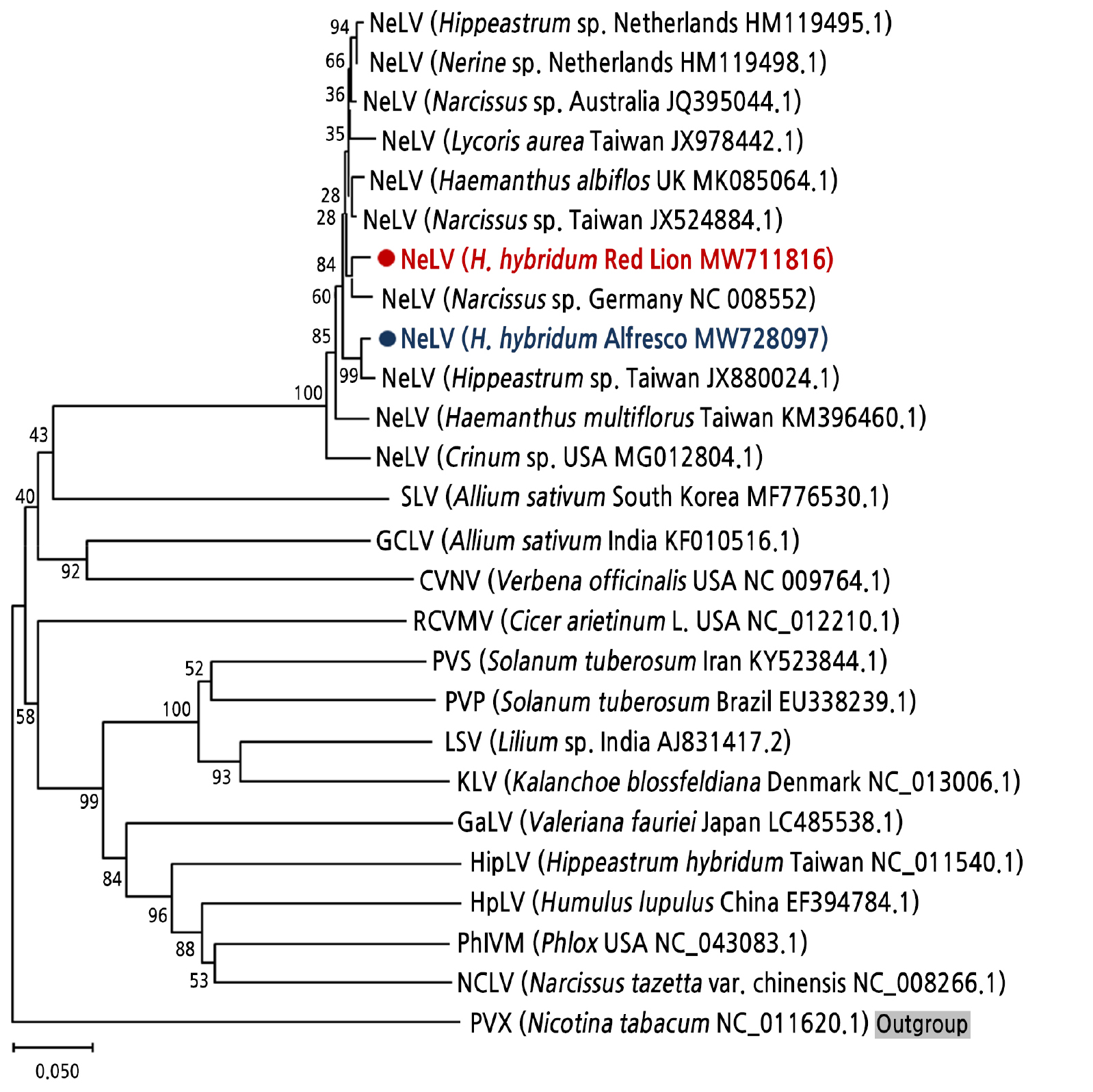
Fig. 2.
Phylogenetic tree constructed using the neighbor-joining method with 1,000 bootstraps based on the partial sequences of the NeLV coat protein gene isolated from the bulb tissues of Hippeastrum hybridum, including nine NeLV sequences, other carlavirus sequences, and an outgroup from NCBI. The following virus species were used in the analysis: Coleus vein necrosis virus (CVNV), Gaillardia latent virus (GaLV), Garlic common latent Virus (GCLV), Hippeastrum latent virus (HipLV), Hop latent virus (HpLV), Kalanchoe latent virus (KLV), Lily symptomless virus (LSV), Narcissus common latent virus (NCLV), Nerine latent virus (NeLV), Phlox virus M (PhlVM), Potato virus P (PVP), Potato virus S (PVS), Potato virus X (PVX), Red clover vein mosaic virus (RCVMV), and Shallot latent virus (SLV).
Discussion
Viruses infecting vegetatively propagated plant species are commonly traded internationally. This study investigated the existence of NeLV in the ornamental bulb Hippeastrum as a starting material for cultivation and trade in domestic and international markets. Studies on NeLV infection have usually used leaf materials for virus detection (Pearson et al., 2009; Jordan et al., 2018) and not the bulb itself. However, leaf materials during growth and cultivation tend to be infected with viruses transmitted by insect vectors, such as aphids (Vijeth et al., 2018). Therefore, the accurate evaluation of the bulb as a starting material for cultivation, whether viruses are present or not, is an important checkpoint. NeLV isolates represented in NeLV are reported to be asymptomatic in the host species of Hippeastrum hybridum, but mosaic symptoms in Crinum (Jordan et al., 2018) and mild streaking in Haemanthus (Monger and Eden, 2019) have been reported. Even though the leaves of amaryllis are asymptomatic, NeLV-infected amaryllis can be a virus source and can transfer this virus to other species causing diseases. In this study, amaryllis bulbs were investigated using RT-PCR to detect NeLV, which has been reported to infect diverse species of Amaryllidaceae. Hippeastrum species have also been reported to be infected with other viruses, such as CMV, HiMV, NYSV, and tobacco rattle virus. In future studies, co-infection needs to be investigated in terms of virome levels through next-generation sequencing. This study describes the results of NeLV investigation and the presence of the virus in an important genus of flower bulbs. To the best of our knowledge, this is the first study to report NeLV infection in Hippeastrum bulb tissue in Korea. As quarantine processes for ornamental bulbous plants are associated with the bulb itself, the information produced from this research will facilitate the accurate identification of viruses, such as NeLV, during cultivation and propagation using a simple RT-PCR-based detection tool.


