Introduction
Materials and Methods
Plant Samples
Chromosome Spread Preparation
FISH
Results
Chromosome Numbers, Length, and Type
Chromosomal Location of Ribosomal DNA and Telomeric Repeat Sequences
Discussion
Introduction
Chromosomal modifications involving alterations in the number and structure of chromosomes are fundamental biological processes that often accompany speciation events; signal the occurrence of genetic diversification; and contribute to the cytological basis of ecological differentiation, adaptation, and reproductive isolation (Madlung, 2013; De Storme and Mason, 2014). The genus Senna, belonging to the subfamily Caesalpiniaceae, provides a pertinent setting for investigating these processes. Encompassing approximately 350 herb, shrub, and tree species, some of which exhibit pharmacological activities and some having found commercial use (Eldemerdash et al., 2022), the Senna genus displays not only broad morphological diversity, including variations in the leaf size and spatial distribution (Oladeji et al., 2021), but also distinct floral morphologies and extrafloral nectaries (EFNs) (Marazzi et al., 2006). The diverse characteristics of Senna plants have motivated cytologists to explore their potential.
The genus Senna displays remarkable variability in its chromosomal number. Previous studies have reported variations in the chromosome number in Senna species, including 2n = 24, 26, 28, 52, and 56 (Irwin and Turner, 1960; Rice et al., 2015; Nguyen et al., 2021). Polyploidy has been observed in S. rugosa with 2n = 42, 56, and 112 (Resende et al., 2014), whereas descending dysploidy has been exemplified by different basic chromosome numbers ranging from x = 11 to x = 14 (Souza and Benko-Iseppon, 2004). In addition, the series Aphyllae exhibited three cytotypes, 2n = 24, 48, and 60, in S. aphylla, whereas S. pachyrrhiza displayed two cytotypes, 2n = 24 and 48 (Robbiati et al., 2022). The most common diploid chromosome number in Senna species is 2n = 28, with a basic chromosome number of x = 14 (Pellerin et al., 2019).
Alterations in repetitive DNA can lead to chromosomal rearrangements and subsequent changes in repetitive DNA through concerted evolutionary mechanisms (Louzada et al., 2020). Chromosomal markers, specifically the mobility of rDNA clusters, provide insight into microevolutionary genomic changes (Raskina et al., 2004). The 5S and 45S rDNA repeat sequences encode ribosomal RNAs essential for ribosomal function and protein synthesis, whereas telomeric repeats can reveal the evolutionary aspects of specific plant species (Waminal et al., 2018). Fluorescence in situ hybridization (FISH) is a valuable technique for deciphering genome structures, compositions, and chromosomal evolution, particularly in closely related species (Waminal et al., 2021).
FISH mapping studies have visualized the chromosomal structures of twenty-three Senna species using 5S and 45S rDNAs and telomeric repeat probes (Youn and Kim, 2018; Pellerin et al., 2019; Nguyen et al., 2021). Nine S. tora repeat-specific pre-labelled oligo probes (PLOPs) were designed and used to observe the chromosomal distribution among the nine Senna species (Ta et al., 2021; Waminal et al., 2021). However, all of these studies focused on diploid Senna species. Although Robbiati et al. (2022) undertook the FISH mapping of the tetraploid S. crassiramea species, their findings did not clearly elucidate the variation in both the chromosome number and structure.
FISH was performed using ribosomal DNA (5S and 45S rDNAs) and telomeric repeat probes to compare the chromosomal compositions and structures of four Senna species: S. artemisioides, S. pallida, S. obtusifolia, and S. aphylla. The results demonstrate the first utilization of triple-color FISH to reveal variations in both the chromosome number and structure in these four species, offering an important reference for understanding chromosomal evolution in the genus Senna.
Materials and Methods
Plant Samples
Accession numbers of seeds of S. artemisioides (DLEG 910206), S. pallida (DLEG 920148), S. obtusifolia (DLEG 910152) and S. aphylla (DLEG 910486) were obtained from the National Plant Germplasm System of the United States Department of Agriculture (USDA). Based on accession numbers, these seeds were collected from different regions, specifically Australia, the United States, Peru, and Argentina (Suppl. Table 1). Following a method in Baskin et al. (1998), the seeds were subjected to a 60 min treatment with sulfuric acid (Sigma-Aldrich, MO, USA) to overcome dormancy, followed by germination in a Petri dish at 25°C. Root tips roughly 2 cm long were initially gathered, treated with 2 mM 8-hydroxyquinoline for 4 h at 18°C, and fixed with a 1:3 solution of glacial acetic acid and absolute ethanol for 24 h. The tips were soaked in 70% ethanol at 4°C for preservation.
Chromosome Spread Preparation
Chromosomal spreads were prepared as described by Waminal et al. (2012). Meristematic root tips (∼2 mm) were collected and then digested by treating them with a 1% concentration of Pectolyase Y-23 (Duchefa Biochemie, Haarlem, Netherlands) and Cellulase R-10 at a concentration of 2% (Duchefa Biochemie) at 37°C for 60 min. After the enzymatic treatment, a 1:3 solution of glacial acetic acid and absolute ethanol was used to re-suspend the pellets, and this was centrifuged at 10,770 × g and at room temperature 20–25°C (RT) for 3 min. The resulting pellet was re-suspended in a 9:1 solution of glacial acetic acid and absolute ethanol. A pipette was used to spread the pellets on pre-warmed glass slides in a humid chamber at 80°C, followed by air drying.
FISH
The universal pre-labeled oligomer probes (PLOPs) for the 5S rDNA, 45S rDNA, and Arabidopsis-type telomeric repeat (TTTAGGG)n developed by Waminal et al. (2018) were used for FISH. PLOPs, typically 20–30 nucleotides long, have a specific design that ensures complementarity with a targeted sequence. This design enhances specificity, thereby augmenting the cost-effectiveness and time-efficiency of a FISH analysis (Waminal et al., 2018).
A hybridization mixture (40 µL) comprising a 50% formamide solution, a 10% solution of dextran sulfate, 2× saline sodium citrate (SSC), at Sigma water at 50 nanograms per microliter for each probe was applied to each slide. The denaturation process was carried out by exposing the slides to a temperature of 80°C for 5 min and incubating them overnight in a moist chamber at RT. Following hybridization, a series of wash steps was conducted on the slides with 2× SSC at RT for 10 min, 0.1× SSC buffer for 25 min at 42°C, and 2× SSC buffer at RT for 5 min, after which they were dehydrated by immersion in a series of ethanol solutions with increasing concentrations of 70%, 90%, and 100% for 3 min each under shaking. Next, 40 µL of a Vectashield (Vector Laboratories, CA, USA)-based solution that contained 4',6-diamidino-2-phenylindole (DAPI) was used on each slide. A BX53 fluorescence microscope (Olympus, Tokyo, Japan) coupled with a DFC365 FS CCD camera (Leica Microsystems, Wetzlar, Germany) and Cytovision ver. 7.2 software (Leica Microsystems) was used to observe and capture the chromosomal spreads.
The best mitotic metaphase spread for each species was selected and analyzed for the chromosome length and type. Chromosome lengths were measured using ImageJ software ver. 1.53 k (Wayne Rasband and contributors, National Institutes of Health, MD, USA), and the images were edited using Photoshop CS6 (Adobe Inc., San Jose, CA, USA). Chromosome pairing was performed based on the ribosomal DNA and telomeric signals, the chromosome length, and the centromere position according to Nguyen et al. (2021).
Results
Chromosome Numbers, Length, and Type
The four Senna species S. obtusifolia, S. pallida, S. aphylla, and S. artemisioides exhibited distinct chromosomal numbers (2n = 26, 28, 48, and 56, respectively; Fig. 1A and Table 1). The basic chromosome number x = 14 corresponded to the tetraploid S. artemisioides (2n = 4x = 56) and diploid S. pallida (2n = 2x = 28), whereas x = 13 and x = 12 corresponded to the diploid S. obtusifolia (2n = 2x = 26) and tetraploid S. aphylla (2n = 4x = 48), respectively. Distinct satellite (SAT) chromosomes were also identified in specific chromosomes (Fig. 1B). Eight SAT chromosomes were observed on chromosomes 5 and 11 in S. artemisioides, whereas S. pallida displayed four on chromosomes 7 and 8, and S. aphylla exhibited four on chromosome 8. S. obtusifolia possesses two SAT chromosomes on chromosome 13.
Table 1.
FISH Karyotype Analyses of the Mitotic Metaphase Chromosomes of Four Senna species
| Species | Chr. no. | Ploid |
Chr. length (µm) |
Karyotypic formular | rDNA signals | Telomeric signals | ||
| 2n | x | 5S | 45S | |||||
| S. artemisioides | 56 | 14 | tetraploid | 1.10–2.83 | 11 mz + 3 smy | 4x (13)w | 8 (5, 11) | Telv |
| S. pallida | 28 | 14 | diploid | 1.72–2.85 | 9 m + 5 sm | 2 (12) | 4 (7, 8) | Tel, ITRu |
| S. obtusifolia | 26 | 13 | diploid | 1.33–3.20 | 13 m | 2 (9) | 2 (13) | Tel, IRst, pCens |
| S. aphylla | 48 | 12 | tetraploid | 0.91–2.68 | 10 m + 2 sm | 4 (12) | 4 (8) | Tel |
r Karyotype assigned according to a method previously reported by Levan et al. (1964).
The ranges of chromosome lengths among the four Senna species are shown in Table 1. The diploid S. obtusifolia exhibited the longest chromosome length, measuring 3.20 µm, whereas the tetraploid S. aphylla displayed the shortest length at 0.91 µm. In S. aphylla, the chromosome lengths ranged from 0.91 µm to 2.68 µm, whereas S. obtusifolia exhibited lengths varying from 1.33 µm to 3.20 µm. The chromosome length range for S. artemisioides was 1.10 µm to 2.83 µm, whereas S. pallida displayed a range of 1.72 µm to 2.85 µm (Suppl. Table 2 and Suppl. Table 3). Based on arm ratios, S. obtusifolia exclusively presented with metacentric chromosomes (Suppl. Table 4). In addition, S. artemisioides exhibited 11 metacentric and three submetacentric chromosomes, S. pallida displayed nine metacentric and five submetacentric chromosomes, and S. aphylla possessed ten metacentric and two submetacentric chromosomes.
Chromosomal Location of Ribosomal DNA and Telomeric Repeat Sequences
The FISH analysis revealed notable variations in the number and position of rDNA and the telomeric signals (Fig. 1). The number of 5S rDNA signals exhibited relatively consistent patterns across the diploid species (S. pallida and S. obtusifolia), with two signals on each pair of chromosomes, whereas the tetraploid species (S. artemisioides and S. aphylla) consistently exhibited four signals. However, substantial discrepancies were observed at specific locations of the signals. In S. pallida and S. aphylla, the 5S rDNA signals were localized in the paracentromeric or centromeric regions of the short arm of chromosome 12, whereas S. artemisioides (chromosome 13) and S. obtusifolia (chromosome 9) showed these signals in the subtelomeric regions of the short arm.
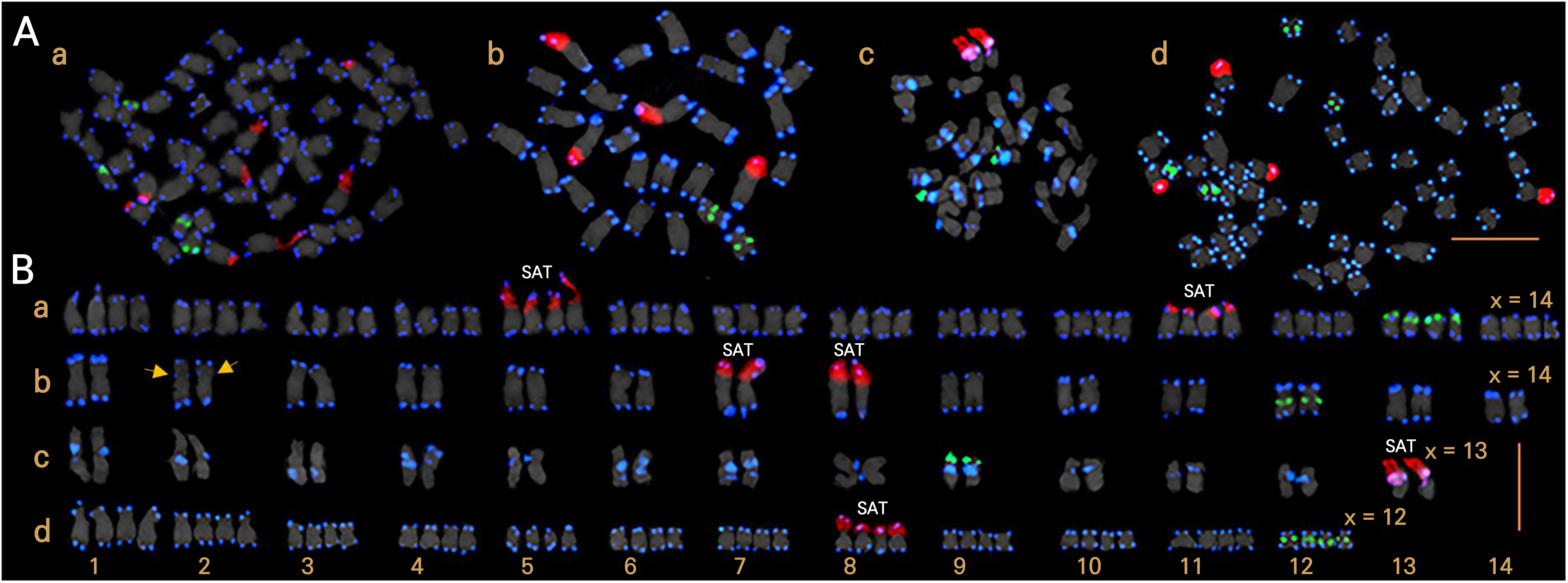
Fig. 1.
FISH analysis demonstrating the metaphase chromosome spreads (A) and karyograms (B) of the four Senna species S. artemisioides (a), S. pallida (b), S. obtusifolia (c), and S. aphylla (d). Satellite (SAT) chromosomes and ITR signals (yellow arrows) were observed. The red, green, and blue signals indicate 45S rDNA, 5S rDNA, and telomeric repeats, respectively. The scale bar is 5 μm.
Significant variations were observed in the number of 45S rDNA signals across the four Senna species, ranging from two to eight. These signals were consistently detected in the nucleolus organizer region (NOR) of the short arm. A minimal number of signals, specifically two, were localized in the NOR of the short arm on chromosome 13 of S. obtusifolia. In contrast, the tetraploid S. artemisioides demonstrated the maximum occurrence of eight signals which were distributed within the NOR of the short arms on chromosomes 5 and 11. S. pallida displayed four signals that were split between chromosomes 7 and 8, which were positioned in the NOR of the short arm. Similarly, S. aphylla showed four signals on chromosome 8, located in the NOR of the short arm.
Notably, significant disparities in telomeric location were particularly prominent in S. obtusifolia, setting it apart from the other three species. In S. artemisioides, S. pallida, and S. aphylla, telomeric signals were primarily observed in the terminal regions of all chromosomes, whereas S. obtusifolia displayed telomeric signals not only in the terminal regions, but also in the interstitial and pericentromeric regions. Specifically, chromosome 4 exhibited a solitary terminal telomeric signal, whereas the remaining chromosomes predominantly showcased telomeric signals in the interstitial and pericentromeric regions. Furthermore, faint interstitial telomeric repeat (ITR) signals were discernible on chromosome 2 of S. pallida.
Discussion
The chromosomal variation, encompassing dysploidy and polyploidy with chromosome numbers of 2n = 26, 28, 48, and 56, observed in the four Senna species investigated here was consistent with previous reports (Souza and Benko-Iseppon, 2004; Rice et al., 2015; Robbiati et al., 2022). However, our study revealed an intriguing correlation between the chromosomal variation in this case and rearrangements of ribosomal DNA and telomeric repeat sites in Senna.
Dysploidy refers to the gradual change in the haploid chromosome count observed in closely related species, leading to the formation of a dysploid series (Guerra, 2008). Among the cytogenetic studies of Senna, the most frequently observed basic chromosome number was x = 14. The primary basic number, x = 7, which was presumed to be the ancestral basic number, established a secondary basic chromosome number, x = 14 (Irwin and Turner, 1960). Earlier work by Souza and Benko-Iseppon (2004) indicated that the basic chromosome number series, i.e., x = 11, 12, 13, and 14, demonstrated a dysploid tendency.
Descending dysploidy mechanisms can result in ITR, which can serve as cytological markers of chromosomal rearrangements, including chromosomal insertions (NCIs) and end-to-end translocations (EETs) (Waminal et al., 2021). ITR signals were frequently present in some diploid Senna species, with x = 14. We observed the presence of ITR signals in S. pallida, and a similar observation was made by Nguyen et al. (2021) in S. alexandrina, S. corymbosa, and S. hirsuta, which supports the notion that x = 14 is a derived number and that other basic numbers, specifically x = 11, 12, and 13, suggest the occurrence of dysploidy in the Senna genus (Waminal et al., 2021).
Polyploidy, the predominant chromosomal variation in plants, involves duplication or multiplication of a complete set of chromosomes. This is the most common chromosomal mechanism driving evolutionary processes in plants (Guerra, 2008). During sexual polyploidization, meiotic restitution or non-reduction results in unreduced or 2n gametes, respectively, and tetraploid individuals are born by the fusion between two diploid gametes (Brownfield and Köhler, 2011). Tetraploid cytotypes exhibit twice the chromosomal complement size of diploid cytotypes (Robbiati et al., 2022). In the genus Senna, based on 5S rDNA signals, this may reflect the ploidy state. Two 5S rDNA signals were detected in all diploids, whereas tetraploids had four signals. However, an increase in 5S rDNA signals was observed in two diploid Senna species with x = 12: S. pachyrrhiza and S. uniflora, each exhibiting three to four chromosomes bearing 5S rDNA signals, (Nguyen et al., 2021; Robbiati et al., 2022).
Furthermore, the variation trend in the 5S rDNA positions among these four species was consistent with a previous study that used three chloroplast regions (rpS16, rpL16, and matK) to establish phylogenetic relationships among ninety-eight Senna species and to gain novel insights into the evolution of EFNs and floral symmetry (Marazzi et al., 2006). Among the seven major clades depicted in the phylogenetic tree, S. artemisioides and S. obtusifolia were grouped into clade IV, whereas S. aphylla and S. pallida were grouped into clade VI. Clade IV consisted of herbs, shrubs, and treelets that possessed EFNs on the leaves and asymmetric flowers, including the corolla, androecium, and gynoecium. Some species exhibit an oblique floral position, enantiostyly, such as S. obtusifolia, and hairs on the anthers of middle and abaxial stamens, which may represent a synapomorphy for clade IV, distinguishing it from other clades. Senna species of the series Interglandulosae, such as S. pallida, are mainly found in moist areas, whereas Aphyllae members, such as S. aphylla, inhabit dry regions. Some species of the series Interglandulosae have up to 14 leaflet pairs, reduced upper petals, and long-beaked anthers. Aphyllae species have photosynthetic stems, few leaves, and unscaled upper petals (Irwin and Turner, 1960; Marazzi et al., 2006).
The two diploid Senna species (S. obtusifolia, S. tora) with x = 13 display a distinct feature that sets them apart from other species: the telomeric signal is located in the terminal, interstitial, and pericentromeric regions instead of only in the terminal regions. Altered telomeric repeat positions were observed in S. obtusifolia for the first time in this study, and Pellerin et al. (2019) reported similar observations in S. tora. EETs may be responsible for the appearance of telomeric signals within interstitial and pericentromeric regions in S. tora were proposed by Waminal et al. (2021). Recent reports have suggested that chromosomal rearrangements (inversions, deletions, duplications, and translocations) are associated with repetitive DNA sequences in heterochromatic regions and heterochromatin-euchromatin borders (Badaeva et al., 2007). Both S. obtusifolia and S. tora are members of the section Prososperma, series Torae (Irwin and Turner, 1960). Based on biogeographic evidence, Randell (1995) proposed that S. tora was derived from S. obtusifolia. The FISH analysis revealed that S. obtusifolia and S. tora exhibit remarkably similar chromosomal numbers, karyotypes, rDNA, and telomeric repeat locations.
The chromosomal structure is strongly affected by ploidy changes and chromosomal rearrangements, which may lead to speciation (Madlung, 2013; De Storme and Mason, 2014). In the genus Carex, dysploidy plays a significant role in speciation (Lipnerová et al., 2013). Chromosomal rearrangements directly affect speciation via reproductive isolation. Two models, the ‘suppressed recombination’ and ‘hybrid sterility’ models, propose the role of chromosome rearrangement in speciation (Faria and Navarro, 2010). In the ‘suppressed recombination’ model, the suppression of recombination over inversion regions facilitates the accumulation of locally adapted alleles, aiding genetic differentiation among overlapping populations (Kirkpatrick and Barton, 2006). The ‘hybrid sterility’ model suggests that reduced fertility in individuals heterozygous for chromosomal rearrangements leads to reproductive isolation of overlapping populations (Ayala and Coluzzi, 2005). In this study, four Senna species were collected from different countries, revealing variations in the chromosome number and structure. These variations suggest that changes in ploidy and chromosome rearrangements have a significant effect on reproductive isolation, driving adaptation and speciation within the genus Senna.
The variation in the chromosome number and structure among the four Senna species (S. artemisioides, S. pallida, S. obtusifolia, and S. aphylla) was visualized by FISH, focusing on rDNA and telomeric repeat probes in an effort to provide potential referral sources for understanding the role of chromosomal changes in species diversification. Additionally, the cytogenetic data presented in this study could be valuable for breeding studies of Senna species. Future studies, including genome size measurements and chromosome painting, will provide further insights into the ploidy state and specific mechanisms underlying genetic rearrangements and will ultimately contribute to a more comprehensive understanding of speciation and evolutionary processes in Senna species.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20240015_Table_1s.pdf
Supplementary Table 1. List of four Senna species and their published chromosome information
- HORT_20240015_Table_2s.pdf
Supplementary Table 2. Karyotype analysis of the best mitotic metaphase chromosome spread in tetraploid S. artemisioides (2n = 56)
- HORT_20240015_Table_3s.pdf
Supplementary Table 3. Karyotype analysis of the best mitotic metaphase chromosome spread in S. pallida (2n = 28)
- HORT_20240015_Table_4s.pdf
Supplementary Table 4. Karyotype analysis of the best mitotic metaphase chromosomal spread in S. obtusifolia (2n = 26)
- HORT_20240015_Table_5s.pdf
Supplementary Table 5. Karyotype analysis of the best mitotic metaphase chromosome spread in tetraploid S. aphylla (2n = 48)


