Introduction
Materials and Methods
Materials and Growth Conditions
Extraction and Determination of Anthocyanin
Extraction of RNA and Genomic DNA
Isolation and Structure Analysis of the Gene and Promoter of IbbHLH2
Construction of Expression Vectors
Stable Expression and GUS Assays
Real-time Quantitative PCR
Results
Isolation and Functional Analysis of the Gene Sequence of IbbHLH2
Isolation and Functional Analysis of the Promoter of IbbHLH2
The Expression Characteristics of IbbHLH2
Discussion
Introduction
Sweet potato [Ipomoea batatas (L.) Lam.] is the sixth largest crop in the world with an annual yield of more than 100 million tons (Loebenstein et al., 2009). Purple-fleshed sweet potato are rich in anthocyanins which can antitumor, antibacterial, antioxidant activities and so on (Springob et al., 2003; Philpott et al., 2004; Yang et al., 2006; Pu and Fu, 2010). Anthocyanins extracted from purple-fleshed sweet potato have a bright color, excellent thermal and light stabilities that can be used to produce high quality anthocyanins.
The synthesis of anthocyanins was regulated by the combination of transcription factors R2R3-MYB, bHLH and WD40 in different plant tissues and developmental stages (Koes et al., 2005; Ramsay and Glover 2005; Lepiniec et al., 2006; Xu et al., 2018). The transcription complex regulated the expression of anthocyanin synthetic genes that control the pattern of accumulation of anthocyanins in plants (Atchley et al., 1999). In most species, the co-expression of R2R3-MYB, bHLH and WD40 strongly induces anthocyanin production (Naing et al., 2018). Six MYB10-like and three bHLH-like TFs were identified as candidates to be the regulators of the anthocyanin accumulation in peach (Rahim et al., 2014).
The transcription factors basic helix-loop-helix (bHLH) are named after their conserved structural domain and typically regulate gene transcription by forming homodimers or heterodimers that bind to target gene promoters (Heim et al., 2003; Holton and Cornish 1995). The best-characterized regulatory genes involved in anthocyanin production were the maize (Zea mays L.) leaf color (Lc) and colorless (CL) genes, which play a role in regulating structural gene expression in maize. The Lc gene strongly activated dihydroflavonol reductase (DFR), flavonoid-3'5'-hydroxylase (F3'5'H), anthocyanidin synthase (ANS) and UDPglucose flavoniod-3-glucosyltransferase (UF3GT) and plays an important role in the anthocyanin synthesis pathway (Bradley et al., 1998). Following the cloning of the maize Lc gene, which encodes a bHLH protein, corresponding genes were found in other plants, such as the Delila gene in snapdragon (Antirrhinummajus) (Naing et al., 2018). In petunia (Petunia hybrida), two related bHLH gene, AN1 and B-Peru regulated the color of the corolla gene (Spelt et al., 2000; Trinh et al., 2016). Glabrous 3(SmGL3) was associated with anthocyanin synthesis in eggplant roots, stems, leaves, flowers, peels and pulp, while the transparent testa (SmTT8) regulated key enzymes downstream of the anthocyanin synthesis pathway (Park et al., 2007). In the Convolvulaceae plant Ipomoea purpurea, the transcriptional regulatory gene IpIVS (IpbHLH2) regulated the expression of related genes in the anthocyanin synthesis pathway, in particular DFR-B and ANS. Transcript levels of DFR-B and ANS were almost zero in the seed coat of plants that did not express IpIVS (Mano et al., 2007). bHLH proteins have also been found to be another key anthocyanin regulator in berry skins (Liu et al., 2015).
The tuberous roots of purple-fleshed sweet potato are rich in anthocyanins. The transcription factors and their regulatory targets involved in the regulation of anthocyanin synthesis in tuberous roots were not well understood. The R2R3 MYB transcription factor gene IbMYB1 had been cloned in purple-fleshed sweet potato. It can regulate the expression of the chalcone synthase (CHS), chalcone isomerase (CHI), F3H, DFR, ANS and 3-glucosyltransferase (3GT) involved in anthocyanin synthesis in purple-fleshed sweet potato (Liu and Chen, 2007). However, it was not clear whether bHLH transcription factor involved in the regulation of anthocyanin synthesis in purple-fleshed sweet potato. In this work, molecular cloning and functional analysis of the IbbHLH2 (GenBank ID: JF508437) to regulation of anthocyanin synthesis in purple-fleshed sweet potato were investigated. The results showed that the color of the cotyledon and epicotyl of IbbHLH2 transgenic plants were purple, their anthocyanin content was increased and the expression levels of anthocyanin-related structural genes (CHS, CHI, F3H, DFR, ANS and 3GT) were up-regulated.
Materials and Methods
Materials and Growth Conditions
Purple-fleshed sweet potato cv. ‘Yamakawamurasaki’ , ‘A5’ , ‘A6’ and white-fleshed sweet potato cv. ‘Yubeibai’ were grown in the Guangdong Key Lab of Biotechnology for Plant Development. Arabidopsis (Arabidopsis thaliana) ‘Columbia’ was grown in a greenhouse with day/night cycle of 16h/8h at 20 ± 2°C. Wild type tobacco (Nicotiana tabacum) ‘NC89’ was grown in a greenhouse under day/night cycle of 16h/8h photo-period at 28 ± 2°C.
Extraction and Determination of Anthocyanin
Plant material (0.3 g) was added to 1.0 ml of methanol containing 1% hydrochloric acid (w/v), and extracted at 21°C for 18 h with shaking at 150 rpm. The extraction solution was obtained after centrifugation at 21500 × g for 3 min and analyzed spectrophotometrically at 530 nm and 657 nm. The anthocyanin content was calculated using the formula: QAnthocyanin = (A530–0.25 × A657) × M-1, QAnthocyanin represents the relative content of anthocyanins, A530 and A657 represent absorbance at 530 nm and 657 nm respectively, and M represents the fresh weight of plant material. All quantitative tests were repeated three times and the data shown are the average of the three repeats ± standard deviation.
Extraction of RNA and Genomic DNA
Total RNA was extracted using the Rapid Extraction Kit from Beijing Baitek Biotechnology Co. Ltd. (Beijing, China) following the manufacturer’s instructions. The RNA concentration was determined using a Nano Drop ND1000 microassay spectrophotometer, and its purity was analyzed by agarose gel electrophoresis. DNA was extracted using a Plant Genomic DNA Extraction Kit (Tiangen Biotech, China).
Isolation and Structure Analysis of the Gene and Promoter of IbbHLH2
Approximately 2 mg of total RNA was reverse transcribed with oligo (dT) as primer using M-MLV reverse transcriptase (Takara, Japan) according to the manufacturer’s recommendations. The IbbHLH2 conservative fragment sequence was extracted using the PrimeScript® One Step RT-PCR Kit Ver.2 (Takara, Japan) following the manufacturer’s instructions. The 3’-cDNA and 5’-cDNA of IbbHLH2 sequence was extracted by the SMART RACE cDNA amplification instructions. PCR products were analysed on 1% agarose gels, for each reaction product, a single fragment was recovered from gels and purifified using a DNA purifification Kit (Magen, Beijing, China). The fragment was ligated into the plasmid, transformed into Escherichia coli DH5α competent cells (Weidi Shanghai, China), and then sequenced (Sangong, Shanghai, China). The primers used for PCR were listed in Suppl. Table 1.
The IbbHLH2 promoter sequence was cloned by hiTAIL-PCR. The reaction mixture (total volume 20 µL) contained 2.0 µL PCR buffer, 2.0 µL dNTP mix, 0.1 µL Ex Taq, 1 µL tuber DNA, 0.6 µL pIbbHLH2R1, 2.0 µL any one of the LAD primers, and water to 20 µL. Each 25 µL reaction mixture contained 2.5 µL PCR buffer, 2 µL dNTP mix, l µL AC1, 1 µL pIbbHLH2R2/R3, 0.1 µL Ex Taq, and 1.0 µL 10-fold-diluted primary Tail-PCR product, plus water to 25 µL. PCR products were analysed on 1% agarose gels, for each reaction product, a single fragment was recovered from gels and purifified using a DNA purifification Kit (Magen, Beijing, China). The fragment was ligated into the plasmid, transformed into Escherichia coli DH5α competent cells (Weidi Shanghai, China), and then sequenced (Sangong, Shanghai, China). The primers used for hiTAIL-PCR were listed in Suppl. Table 1.
The IbbHLH2 ORF and nucleotide sequences was translated by ORF Finder tool in NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). IbbHLH2 biochemical properties was predicted by ProtParam tool and secondary structure was analysis by SOPMA software in Expasy (http://www.expasy.ch/tools/). Three-dimensional Model of IbbHLH2 was predicted and analysis by Swiss-Model software (http://swissmodel.expasy.org/) and Weblab viewlite 4.0 software. The structure analysis of IbbHLH2 promoter was pridicted by PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and Plant-CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Construction of Expression Vectors
To identify the function of the IbbHLH2 gene of purple-fleshed sweet potato cv. ‘A5’ in the regulation of anthocyanin synthesis, the plant overexpression vector 35S::IbbHLH2 gene was constructed and transformed into the wild type arabidopsis thaliana under the mediation of agrobacterium EHA10. Seeds of T1 generation were harvested and planted on an MS plate containing Hyg antibiotic. DNA was extracted from the growing seedlings and PCR was performed to confirm that the recombinant plasmid had been transferred to arabidopsis thaliana.
The pCambia1300vector was used for plant expression constructs. The PCR product was purified and recovered using AxyPrep DNA Gel Recovery Kit (Axygen) according to the manufacturer’s instructions. The amplified products were analyzed on 0.8% agarose gels, and single fragments were recovered from the gels. pCambia1300 was digested with KpnI and NcoI, the resulting fragment was ligated into the bHLH2 DNA fragment and then transformed into E. coli DH5α. Constructs were confirmed by sequencing. pCambia1300containing IbbHLH2 was transformed into A. tumefaciens strain EHA105.
To examine the functions of the IbbHLH2 promoter, pIbbH2 was fused with the GUS reporter gene in the pBI121 binary vector to generate the plant expression vector pIbbHLH2::GUS. The pIbbHLH2 sequence was amplified from mountain purple genomic DNA using the specific primers pIbbHLH2F and pIbbHLH2R. The recombinant vector was then transformed into E. coli DH5α and identified by PCR. After agarose gel electrophoresis, a target band of about 1000 bp was observed, indicating that the pIbbHLH2::GUS plant overexpression vector had been successfully constructed. The recombinant plasmid pIbbH2::GUS was transformed into Agrobacterium EHA105 and its presence confirmed by PCR. The pIbbHLH2::GUS plasmid was introduced into tobacco leaves by Agrobacterium-mediated transformation, and GUS activity was assessed in infected leaves transiently expressing either pIbbHLH2::GUS or 35S::GUS.
pIbbHLH2 was recombined into the pBI121 vector enzymed by HindIII and BamHI, replacing the 35S promoter sequence. The IbbHLH2 promoter was truncated for construct chimeric genes consisting of GUS driven by various IbbHLH2 promoter regions. Full-length of pIbbHLH2 were fused to a beta-glucuronidase (GUS) reporter gene to investigate the activity of IbbHLH2 promoter. The PCR product was subcloned into the pBI121 vector by replacing the CaMV35S promoter in PBI121. Positive clones were verified by sequencing. The pBI121-bearing was transferred into A. tumefaciens strain EHA105. The primers used for constructing vectors were listed in Suppl. Table 1.
Stable Expression and GUS Assays
Transgenic Arabidopsis plants were generated by the standard floral-dip protocol as described elsewhere (Schoenbohm et al., 2000). Transformants were selected by growing on MS medium containing 50 mg·L-1 kanamycin. All hygromycin- resistant T3 plants were analyzed by PCR. Beta-glucuronidase (GUS) histochemical assays were performed according to the method described as our reported previously (Fu et al., 2020).
Real-time Quantitative PCR
The expression of transcription factors (IbMYB1, IbbHLH2 and IbWD40) in fibrous roots, thick roots and storage roots of purple-fleshed sweet potato cv. A5 and white-fleshed sweet potato cv. Yubeibai were analyzed using real-time quantitative PCR. First-strand cDNA was synthesized from total RNA using Prime Script™ RT Master Mix (Takara). RT-qPCR was conducted using SYBR® Premix Ex Taq™ II (Takara) in a total reaction volume of 20 µL consisting of 100 ng of template cDNA, each primer at 0.5 µM and 10 µL of SYBR® Premix Ex Taq™ II, and the amplification program was as follows: 1 cycle of 95 °C for 10 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s with a Bio-Rad CFX96 Real-Time PCR system (BIO-RAD, USA) according to the manufacturers’ instructions. IbG14 was used as internal controls and calculated using the comparative Ct analysis method. The primers used for RT-qPCR were listed in Suppl. Table 1.
Results
Isolation and Functional Analysis of the Gene Sequence of IbbHLH2
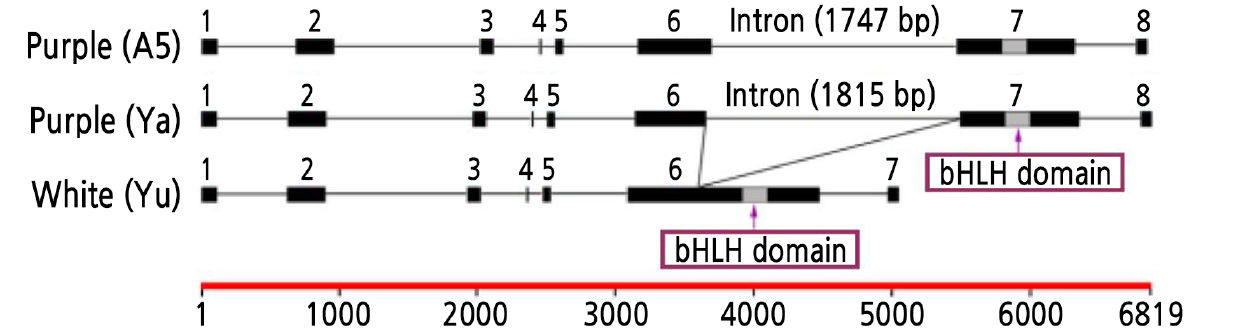
In this study, the ORFs of the IbbHLH2 genes of ‘Yamakawamurasaki’, ‘A5’ and ‘Yubeibai’ were cloned and systematically analyzed. Three fragments with a uniform size of about 2000 bp were obtained from cDNA of the three variety, which were named IbbHLH2-Ya (Yamakawamurasaki), IbbHLH2-A5 and IbbHLH2-Yu (Yubeibai). Sequence analysis showed that the three fragments have 94.8% cDNA sequence identity and 92.0% amino acid sequence identity. The amino acid sequence of the ‘Yubeibai’ had seven amino acid changes compared with the ‘Yamakawamurasaki’ and ‘A5’, but no differences were found in the key functional bHLH domain (Fig. 1).
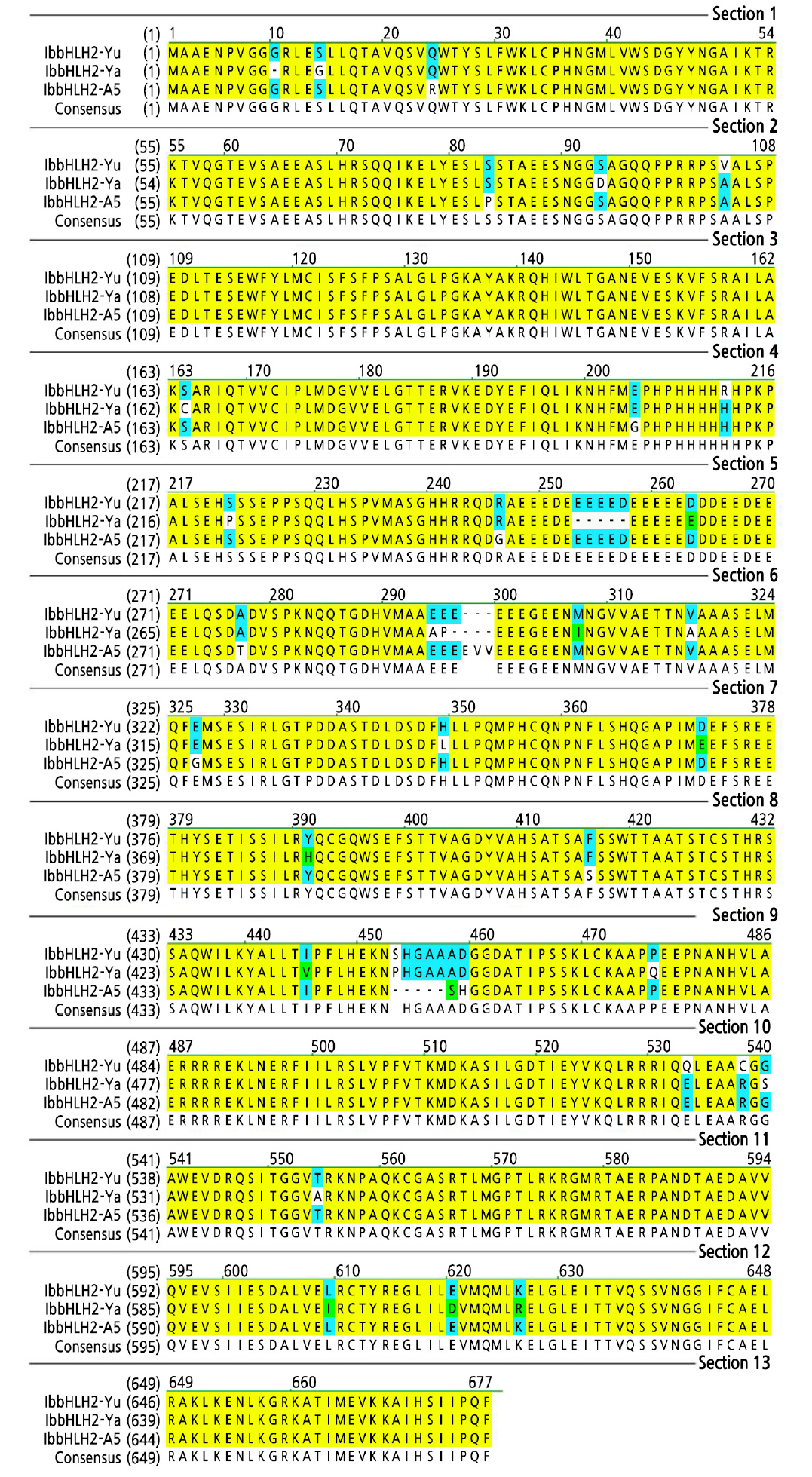
Fig. 1.
Comparison of the amino acid sequences encoded by the IbbHLH2 gene in the purple-fleshed and white-fleshed sweet potato. Ya and A5 was purple-fleshed sweet potato cv. ‘Yamakawamurasaki’ and ‘A5’, Yu was white-fleshed sweet potato cv. ‘Yubeibai’. The green color on the sequence represent the different of the amino acid sequences encoded by the IbbHLH2 gene in the purple-fleshed and white-fleshed sweet potato.
The genomic DNA sequences of the four ORF regions of the IbbHLH2 gene were obtained by PCR amplification using the segmented cloning strategy from the same three variety. After splicing, the complete coding sequences of three variety of IbbHLH2 genomic DNA were obtained and named gIbbHLH2-Ya, gIbbHLH2-A5 and gIbbHLH2-Yu (Fig. 2). gIbbHLH2-Ya was 6819 bp in length and contained eight exons and seven introns and the intron boundary sequences were in line with GT-AG rules. gIbbHLH2-A5 was 6755 bp in length and also contained eight exons and canonical exon boundary sequences. gIbbHLH2-Yu was 4975 bp in length containing seven exons with conventional boundary sequences. It can be seen from the above results that the IbbHLH2 genes of the two purple-fleshed sweet potato have an extra exon and intron compared with the white-fleshed sweet potato. Further analysis revealed that the lengths of the extra intron in the purple-fleshed sweet potato were 1815 bp in gIbbHLH2-Ya and 1747 bp in gIbbHLH2-A5, and the insertion position was in the middle of the sixth exon of the white-fleshed gene, located 323 bp upstream of the bHLH functional domain (Suppl. Fig. 1). Further analysis of the extra intron sequences in gIbbHLH2-Ya and gIbbHLH2-A5 revealed that they were very similar (86.2%), both containing MYB binding sites, multiple photoresponsive elements and hormone (ABA, GA and SA) response elements. Therefore, the results indicated that this intron may regulate the expression of the IbbHLH2 gene in purple-fleshed sweet potato.
To identify the function of the IbbHLH2 gene in the regulation of anthocyanin synthesis, the plant overexpression vector 35S::IbbHLH2 gene was constructed and transformed into the wild type arabidopsis thaliana under the mediation of agrobacterium EHA10. Seeds of T1 generation were harvested and planted on an MS plate containing Hyg antibiotic. DNA was extracted from the growing seedlings and PCR was performed to confirm that the recombinant plasmid had been transferred to arabidopsis thaliana. The T3 generation transgenic homozygous plants were obtained through twice on Hyg resistance screening. Genomic DNA and RNA were extracted from homozygous transgenic lines, and the IbbHLH2 gene sequence of about 2000 bp was obtained by genomic PCR amplification, indicating that the gene IbbHLH2 has been successfully inserted into the wild-type Arabidopsis genome (Fig. 3). Semi-quantitative RT-PCR showed that the IbbHLH2 gene was expressed at high levels in transgenic plants (Fig. 3).
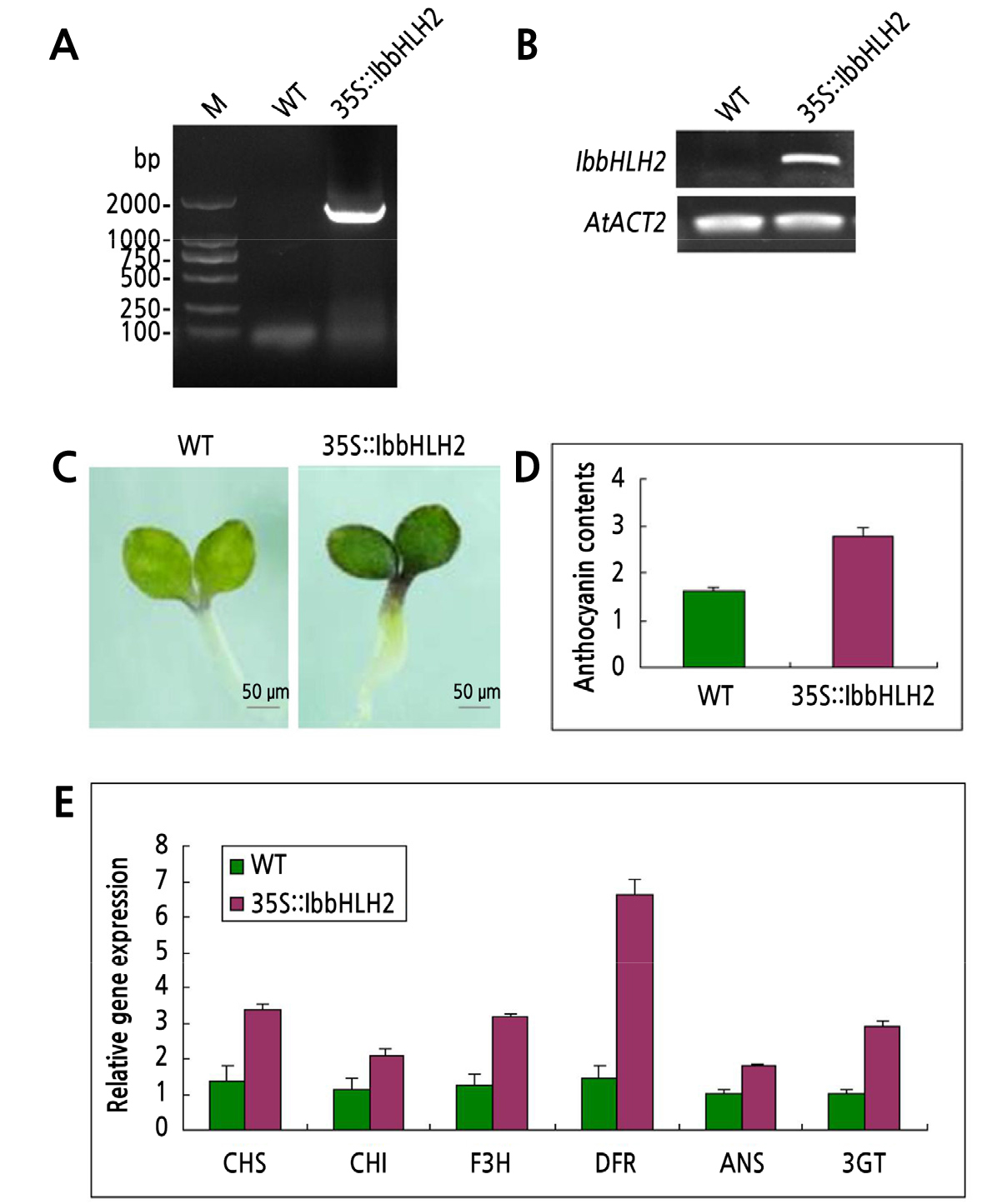
Fig. 3.
Functional verification of IbbHLH2 in transgenic Arabidopsis. A: PCR detection of IbbHLH2 gene insertion in transgenic Arabidopsis genome, B: RT-PCR detection of IbbHLH2 gene expression in transgenic Arabidopsis, C: wild-type and transgenic seedlings, D: content of anthocyanin in flowers of wild-type and transgenic Arabidopsis seedlings, E: expression level of anthocyanin synthesis-related structural genes in wild-type and transgenic Arabidopsis seedlings. The unit of anthocyanin content is g-1FW.
The color traits, anthocyanin content and related structural gene expression were investigated in transgenic A. thaliana(Suppl. Fig. 1). The result showed that the cotyledons and epicotyls of the seedlings of transgenic plants emerge purple color and the anthocyanin content was higher than in controls. Furthermore, the expression levels of anthocyanin synthesis-related structural genes (CHS, CHI, F3H, DFR, ANS and 3GT) were all higher in transgenic plants. This results indicated that the IbbHLH2 gene cloned in this paper was a functional sequence and that its introduction into the Arabidopsis genome can increase the content of anthocyanins in the resultant transgenic plants by up-regulating the expression level of related bio-synthetic genes.
Isolation and Functional Analysis of the Promoter of IbbHLH2
The genomic DNAs of ‘Yamakawamurasaki’, ‘A5’ and ‘Yubeibai’ were used as templates to clone the promoter sequences, which were named pIbbH2-Ya (Yamakawamurasaki), pIbbH2-A5 (A5) and pIbbH2-Yu (Yubeibai). Sequence analysis showed that the above three sequences were very similar with an identity of 99.1% and no base mutations in the positions of the key cis-acting elements (Suppl. Fig. 2).
The promoter of IbbHLH2 (pIbbH2) was obtained from purple-fleshed sweet potato ‘Yamakawamurasaki’ by hi-TAIL PCR and shown by sequencing to be 1020 bp in length (Suppl. Fig. 3). Analysis of the sequence revealed that pIbbH2 contains a number of cis-acting transcription elements, MYB binding elements, a TATA-box, a CAAT-box, a MYC binding site, an abscisic acid (ABA) response element (ABRE), two binding sites for the transcription factor WRK710S associated with the gibberellin (GA) response and WBOX elements associated with the salicylic acid (SA) response. In addition, multiple photo-responsive structural elements were found in pIbbHLH2.
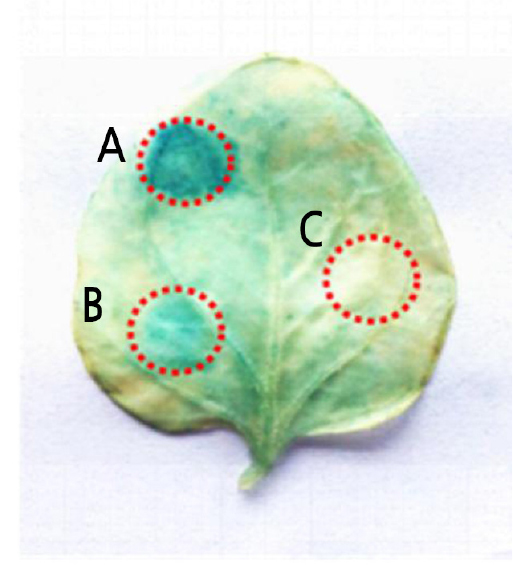
To examine the functions of the IbbHLH2 promoter, pIbbH2 was fused with the GUS reporter gene in the pBI121 binary vector to generate the plant expression vector pIbbHLH2::GUS. The pIbbHLH2 sequence was amplified from mountain purple genomic DNA using the specific primers pIbbHLH2F and pIbbHLH2R. pIbbH2 was recombined into the pBI121 vector enzymed by HindIII and BamHI, replacing the 35S promoter sequence. The recombinant vector was then transformed into E. coli DH5α and identified by PCR. After agarose gel electrophoresis, a target band of about 1000 bp was observed, indicating that the pIbbHLH2::GUS plant overexpression vector had been successfully constructed. The recombinant plasmid pIbbH2::GUS was transformed into Agrobacterium EHA105 and its presence confirmed by PCR. The pIbbHLH2::GUS plasmid was introduced into tobacco leaves by Agrobacterium-mediated transformation, and GUS activity was assessed in infected leaves transiently expressing either pIbbHLH2::GUS or 35S::GUS. The results indicate that pIbbH2 has a promoter activity which can initiate the expression of the GUS gene (Fig. 4).
The Expression Characteristics of IbbHLH2
To explore the relationship between the expression pattern of the IbbHLH2 and anthocyanin biosynthesis in the root of sweet potato, three purple-fleshed sweet potato and one white sweet potato were researched (Fig. 5). The anthocyanin content and the expression levels of IbbHLH2 in different tissues at different developmental stages of the roots were measured. The results showed that the expression of IbbHLH2 was completely synchronized with the accumulation of anthocyanins in different variety and developmental stages.
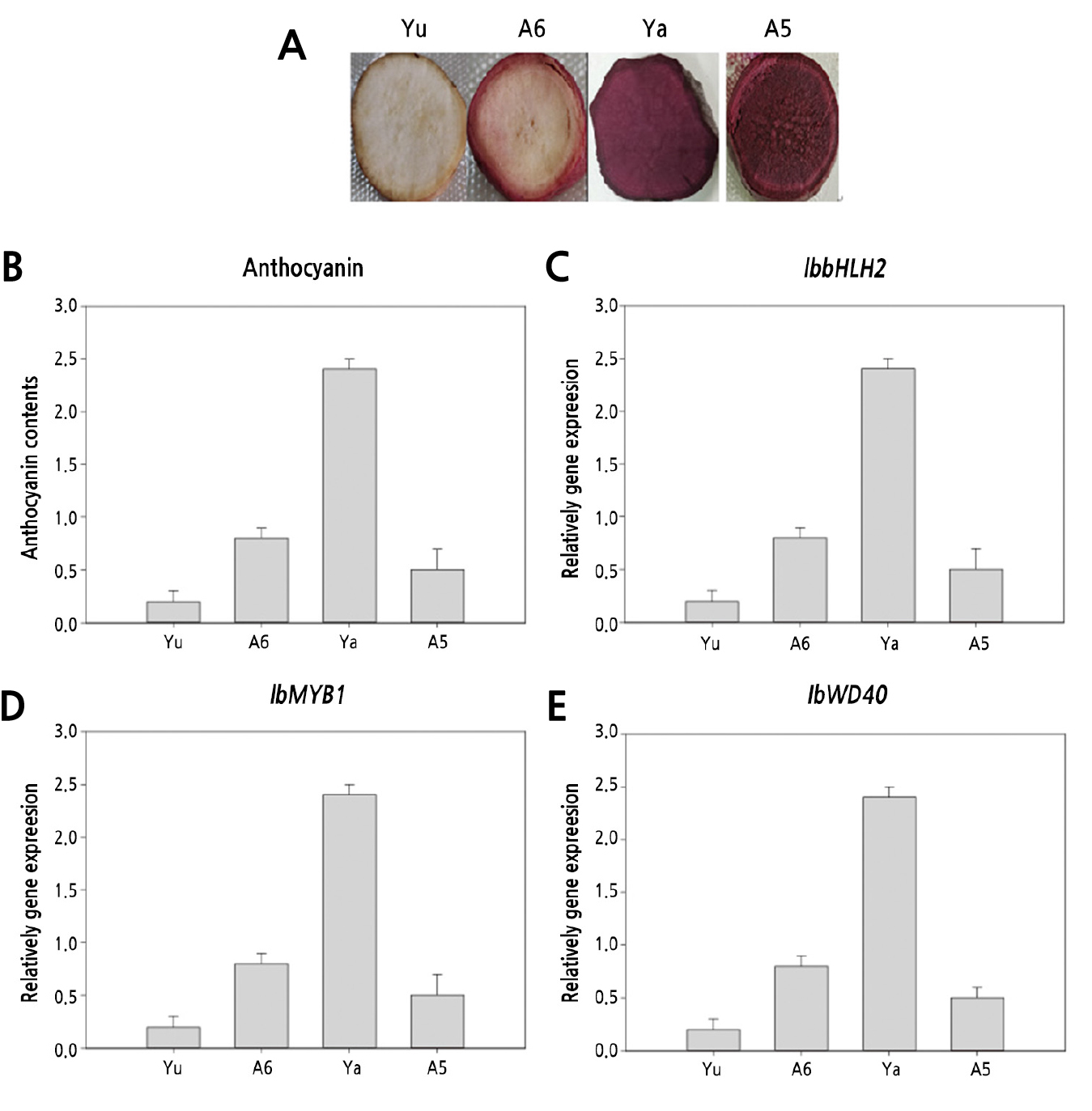
Fig. 5.
Anthocyanin content and expression of transcription factors in different sweet potato. A: Cross-section of sweet potato roots; B: Anthocyanin content in different sweet potato; C, D and E: Expression of transcription factors in different sweet potato. Yu (‘Yubeibai’), Ya (‘Yamakawamurasaki’), A5 (‘A5’), A6 (‘A6’). The unit of anthocyanin content is g-1FW.
The transcription levels of three transcription factor genes (IbbHLH2, IbMYB1 and IbWD40) involved in anthocyanin synthesis were assessed by qRT-PCR in roots of the four sweet potato cultivars. The expression levels of IbbHLH2 and IbMYB1 across the four sweet potato were consistent with anthocyanin content and the expression levels of key anthocyanin biosynthesis pathway genes. IbWD40 represented an exception to this general trend, since its expression level was highest in ‘Yamakawamurasak’ rather than in ‘A5’ (Fig. 6). The result indicated that IbbHLH2 and IbMYB1 were the key transcription factors that regulate the synthesis of anthocyanin in rhizomes of purple sweet potato while IbWD40 may be less important.
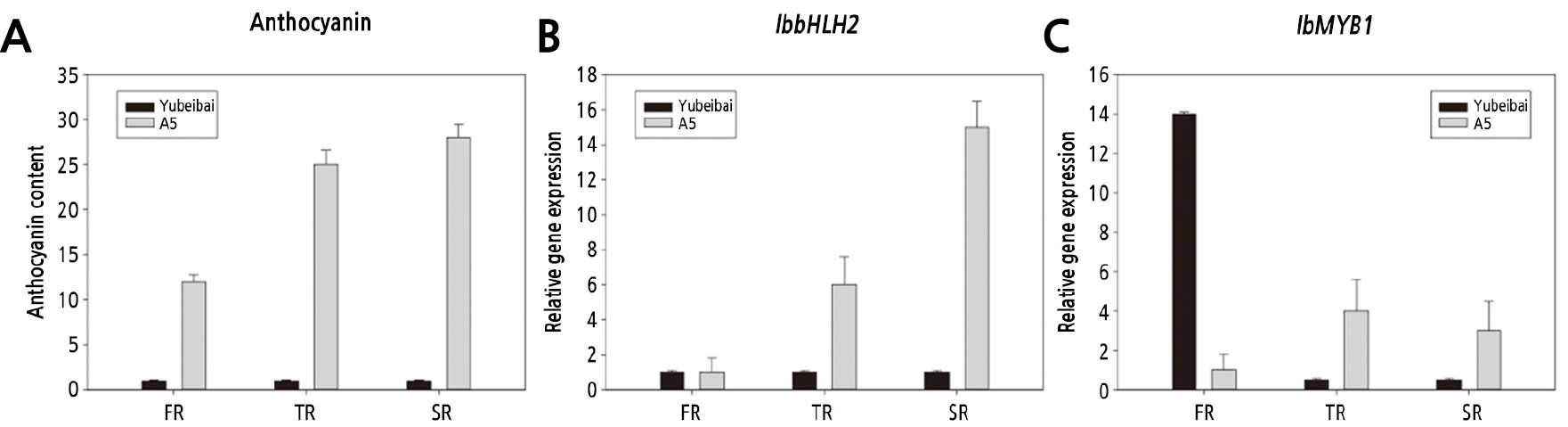
Fig. 6.
Anthocyanin content and expression of transcription factors in different developmental stages of purple-fleshed and white-fleshed sweet potato. FR: Fibrous roots (diameter < 2 mm), TR: Thick roots (2 mm < diameter < 5 mm), SR: Storage roots (diameter > 5 mm). The unit of anthocyanin content is g-1FW.
The anthocyanin content and the expression level of structure genes and transcription factors were measured in purple-fleshed strain ‘A5’ with the highest anthocyanin content and white-fleshed variety ‘Yubeibai’ with the lowest anthocyanin content at different developmental stages. Three developmental stages as foollows: fibrous roots (diameter < 2 mm), thick roots (2 mm < diameter < 5 mm) and storage roots (diameter > 5 mm) .
In the purple-fleshed sweet potato, the accumulation of anthocyanin content gradually increased as the roots developed and was highest in mature roots, the expression pattern of the IbbHLH2 gene was also consistent with the trend in anthocyanin content, namely, fibrous roots < thick roots < storage roots. IbMYB1 showed the highest expression in storage roots, and the lowest level in fibrous roots, which was inconsistent with the trend in anthocyanin content.
In white-fleshed sweet potato, the anthocyanin content was decreased gradually as roots matured (Fig. 4), the expression levels of all the tested genes were consistent with the trend of anthocyanin content, namely, fibrous roots > thick roots > storage roots. The expression levels of all the genes tested and the anthocyanin content in thick roots and mature storage roots of the purple-fleshed was higher in the white-fleshed sweet potato. IbbHLH2 was expressed at a higher level in purple-fleshed roots while IbMYB1 was higher in the white variety at the fibrous root. These results indicated that both IbbHLH2 and IbMYB1 involved in the regulation of anthocyanin synthesis in the thick roots and mature storage roots, while in the fibrous root, IbbHLH2 play a key role in transcriptional regulation at the synthesis of anthocyanins.
In summary, the expression of the IbbHLH2 gene was completely synchronized with the synthesis and accumulation of anthocyanins in different sweet potato variety and their different root developmental stages, indicating that IbbHLH2 is the key transcriptional regulatory gene involved in the synthesis and metabolism of anthocyanins in purple-fleshed sweet potato.
Discussion
The Lc (leaf colour) gene, a member of the maize R gene family, was the first reported plant gene to encode a transcription factor with the bHLH domain. It is crucial for the synthesis of maize anthocyanins. Further studies revealed that Lc interacts with the maize MYB transcription factor C1 to regulate the synthesis of anthocyanins, and that Lc interacts with the structural gene promoter sequence 5’-CAGGTG-3’, which is found in genes encoding enzymes of the anthocyanin synthesis pathway, including the CHS 5’-CACGTA-3’, CHI 5’-CACGTG-3’and DFR 5’-CACGTG-3’ sites (Li et al., 2007). Transformation of the Lc gene into petunia produces a significant increase in expression of PAL, DFR, F3’H, F3H, ANS and UFGT (Bradley et al., 1998). Studies of Lc gene-transformed apples have also shown that PAL, CHS, F3’H, DFR and ANS are upregulated (Aboudi et al., 2012). In this research, the cognate IbbHLH2 gene is expressed coordinately with accumulation of anthocyanin, indicating that IbbHLH2 is the key transcriptional regulatory gene in the synthesis and metabolism of anthocyanins in purple-fleshed sweet potato. The low level of expression of IbbHLH2 in white-fleshed sweet potato is likely one of the main reasons for the lack of color in roots of this variety.
Although the comparison of IbHLH2 ORF sequences were changed in seven amino acid residues, there were no differences in the functional domain. This indicated that the loss of pigment synthesis in the white-fleshed cultivar was not caused by a change in the structure of IbHLH2 protein, and there may be no difference in function between the purple-fleshed and white-fleshed versions of IbbHLH2 protein. However, the genomic DNA sequences of IbbHLH2 in purple and white-fleshed variety had significant differences. An extra intron, located 323 bp upstream of the sequence encoding the bHLH domain, had been inserted into the purple-fleshed sweet potato IbbHLH2 gene. Further preliminary examination of this intron revealed that it contained a large number of cis-acting elements, including MYB binding elements, as well as light and hormone (ABA, GA and SA) response elements. Thus, this intron sequence may had a regulatory effect on the expression of the IbHLH2 gene.
In recent researches, there had been a number of reports of the regulation of gene transcription by introns. In general, the presence of introns in yeast, nematodes, insects, mammals and plants has a positive effect on gene expression (Rose 2002). In plants, a variety of introns had been found to enhance gene expression. For example, A. thalianaUBQ10 intron 1 can increase gene expression levels 15-fold, inclusion of maize shl gene subunit 1 can increase the expression level of a reporter gene by 40-fold, the intron of the SeFAD2 gene of sesame also had an enhanced expression effect (Kim et al., 2006). The regulation of gene expression by introns occurs not only during pre-mRNA splicing, but can also had broader effects on the regulation of gene expression (Maquat and Carmichael 2001; Maniatis and Reed 2002; Orphanides and Reinberg 2002; Dong et al., 2014; Xu et al., 2014). Therefore, the extra intron in IbbHLH2 may enhance the expression of its cognate gene by interacting with transcription factors. The details of this regulatory mechanism will be the focus of further studies. The identification of IbbHLH2 as a key transcriptional regulatory gene in the anabolism of anthocyanin in purple sweet potato lays the foundations for detailed molecular studies of the synthesis and accumulation of anthocyanins in purple sweet potato tuber, and may stimulate new approaches for molecular-oriented breeding of sweet potato pigment traits (anthocyanin content). In this research, the study was based on an indirect functional test in the model system, although the genetic transformation system of I. batatas Lam. was already established. However, there needs more researchs to evidence the main idea in the future.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20220007_Table_1s.pdf
The primer sets used in this research
- HORT_20220007_Figure_1s.pdf
The sequence and motifs of IbbHLH2 in purple-fleshed sweet potato. The red color of the box on the sequence represent the domain of CAAT-box. The yellow color of the box on the sequence represent the domain of ABA. The blue color of the box on the sequence represent the domain of TATA-box. The green color of the box on the sequence represent the domain of MYB binding site. The orange color of the box on the sequence represent the domain of light responsive element.
- HORT_20220007_Figure_2s.pdf
Alignment of the promoter squences of the IbbHLH2 genes from purple-fleshed and white-fleshed sweet potato.
- HORT_20220007_Figure_3s.pdf
The sequence and motifs of pIbbHLH2 in purple-fleshed sweet potato.