Introduction
Materials and Methods
Plant Materials and RNA Extraction
Characterization of VfRING1a
cDNA Synthesis and Real-time PCR Analysis
Results
Identification, Conserved Domains, and Structure Analysis of VfRING1a
Expression Pattern of VfRING1a
Discussion
Introduction
Polycomb group (PcG) proteins are crucial epigenetic regulators, first discovered in Drosophila (Lewis, 1978; Vidal, 2009). PcG proteins were reported to be subunits of multiprotein complexes that exhibit enzymatic activities of specific histone modification (Calonje, 2014). The two best-characterized subunits of PcG present in Drosophila and mammals are polycomb repressive complex1 (PRC1) and PRC2 (Lanzuolo and Orlando, 2012). PRC2 catalyzes histone H3 lysine 27 (H3K27) trimethylation (H3K27me3), which is subsequently recognized by PRC1, which further catalyzes H2A monoubiquitination (H2Aub1), creating a transcriptionally silent chromatin conformation (Molitor and Shen, 2013).
Based on the homology and domain organization, the RING1 (Really Interesting New Gene 1) subfamily in Arabidopsis is composed of AtRING1a and AtRING1b (Sanchez-Pulido et al., 2008; Xu and Shen, 2008). AtRING1a and AtRING1b are identified as the core components of PRC1 RING-finger proteins (Sanchez-Pulido et al., 2008). PRC1-like finger protein activity has also been reported in Arabidopsis (Aubert et al., 2001; Levy et al., 2002).
AtRING1a and AtRING1b play a role in regulating the floral transition and shoot apical meristem activity in Arabidopsis (Xu and Shen, 2008; Shen et al., 2014). RING-finger protein members are widely present in eukaryotic cells and are indicated to have a crucial function in ubiquitin-dependent protein degradation, which functions in E3 ubiquitin ligase activity (Jackson et al., 2000; Lin et al., 2008). The ubiquitination mediates numerous processes in plant development, such as embryogenesis, hormone signaling, and senescence, and also in plant defense response (Takai et al., 2002; Moon et al., 2004; Serrano and Guzmán, 2004; Dreher and Callis, 2007). In this pathway, E3 ubiquitin ligase plays a key role in determining candidate proteins for ubiquitination (Ciechanover, 1998).
In the present study, we identified the VfRING1a gene by comparing with the RING-finger protein gene AtRING1a of Arabidopsis. The expression profile of VfRING1a in different tissues and pathogen-inoculated leaves was analyzed by real-time PCR analysis. The results obtained will be beneficial for future studies of VfRING1a structure and function in grapevines.
Materials and Methods
Plant Materials and RNA Extraction
Vitis flexuosa grapevines were grown at 25°C/18°C (day/night) and 65% relative humidity in an experimental greenhouse at Yeungnam University, Gyeongsan, Korea. Leaves, flowers in prebloom and full bloom stages, and young and ripe berries without visible damage and signs of pathogens were collected from the grapevines. All samples were frozen immediately in liquid nitrogen and stored at ‑ 80°C until further use.
The grapevines (‘Campbell Early’, ‘Kyoho’, and V. flexuosa) were inoculated with the pathogens, Botrytis cinerea, Colletotrichum acutatum, C. gloeosporioides, Elsinoe ampelina, and Rhizobium vitis. The treated plants were placed in a plastic box and kept in the dark at 25°C for 4 d. Leaves were harvested at 0, 1, 6, 24, and 48 h after inoculation, immediately frozen in liquid nitrogen, and stored at ‑ 80°C for future use.
Total RNA was extracted from about 1 ‑ 3 g of each sample at different developmental stages of V. flexuosa, using the pine tree method described by Chang et al. (1993), with slight modifications of centrifugation speed (3,200 rpm). All RNA extractions were performed in triplicate. RNA quantity and quality were determined based on absorbance at 230, 260, and 280 nm, which was measured using a Nabi UV/Vis Nano Spectrophotometer (MicroDigital Co., Ltd., Daejeon, Korea). The integrity was assayed through electrophoresis using a 1.2% agarose gel.
Characterization of VfRING1a
Specific primers for real-time PCR were designed by nucleotide sequences of the differentially expressed VfRING1a genes, which were detected by the transcriptome data of V. flexuosa infected with E. ampelina (Ahn et al., 2014) (Table 1).
Table 1.
Nucleotide sequences of gene primers used for sequencing and quantitative real-time PCR analysis
yNCBI: National Center for Biotechnology Information.
The MEME program (https://meme-suite.org/) was used to analyze the structural distinction among VfRING1a, VvRING1a, and AtRING1a, whereas a SWISS-MODEL protein structure prediction was applied to analyze the 3D steric structure of the RING1a genes (https://swissmodel.expasy.org/).
cDNA Synthesis and Real-time PCR Analysis
First-strand cDNA was synthesized from 500 ‑ 1,000 ng of total RNA using the GoScriptTM Reverse Transcription System (Promega, Madison, WI, USA), after which the obtained samples were used as a template for PCR. Expression analysis of VfRING1a was conducted by quantitative PCR using a C1000TM Thermal Cycler (CFX96TM Real-Time System, BioRad, Foster City, CA, USA) and SYBR Premix Ex (TaKaRa Bio Inc., Osaka, Japan). Amplification was conducted by subjecting the samples to one cycle at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Transcript levels were calculated using the standard-curve method, normalized against the grapevine beta-actin gene (AB372563; Table 1) as an internal control, with subsequent measurement of melting curves of the amplified products. Noninoculated leaves were used as the reference sample, defined as the 1× expression level. Results are expressed as the fold increase in mRNA over the reference sample. All qPCR experiments were performed in triplicate to ensure consistency of the results using gene-specific primers. Gene-specific primer pairs were designed using Primer3 software (http://frodo.wi.mit.edu/primer3) and employed for real-time PCR amplification.
Results
Identification, Conserved Domains, and Structure Analysis of VfRING1a
VfRING1a was selected from the transcriptome analysis data of V. flexuosa leaves inoculated with E. ampelina (Ahn et al., 2014). VvRING1a (accession nos. XM010658841.2, XM010657144.2, and XM010657143.2) and AtRING1a (accession no. NM123795.5) were used for nucleotide sequence analysis using the NCBI BLAST search tool (http://blast.ncbi. nlm.nih.gov/Blast.cgi). The respective specimens were deposited at the National Agricultural Biotechnology Information Center (NABIC), Rural Development Administration, Korea, under accession number NS-2921. The predicted VfRING1a protein comprises 480 amino acids with a molecular weight of 54.491 kD and a pI of 5.69. A RING domain in the N-terminal end and a ubiquitin-like domain at the C-terminal end in VfRING1a were identified by the SMART programs (http://smart.embl-heidelberg.de/). It was determined that VfRING1a has similar amino acid sequences as VvRING1a, all containing the conserved RING domains (Fig. 1). The RING domain of VfRING1a shares 51.3% to 98.3% identity with AtRING1a and VvRING1b, the plant defense-related RING finger proteins, and harbors eight conserved Cys and/or His residues coordinated with two zinc ions that are fundamental for E3 ubiquitin ligase activity (Stone et al., 2005). This indicates that the VfRING1a protein probably plays a role in defense response in grapevines by modifying the proteome via ubiquitination.
The amino acid sequences of five proteins were aligned to determine various motifs using the MEME motif search tool. We found 12, 11, and 8 conserved motifs dispersed throughout the proteins in VfRING1a_Locus13360 (NS-2921, NABIC) and VvRING1a_XM010658841.2, VvRING1a_XM010657144.2 and VvRING1a_XM010657143.2, and AtRING1a_ NM123795.5, respectively (Fig. 2). Motif 1 was found to be related to the RING-type zinc-finger domain. The block diagram and logos of the conserved motifs are presented in Fig. 2.
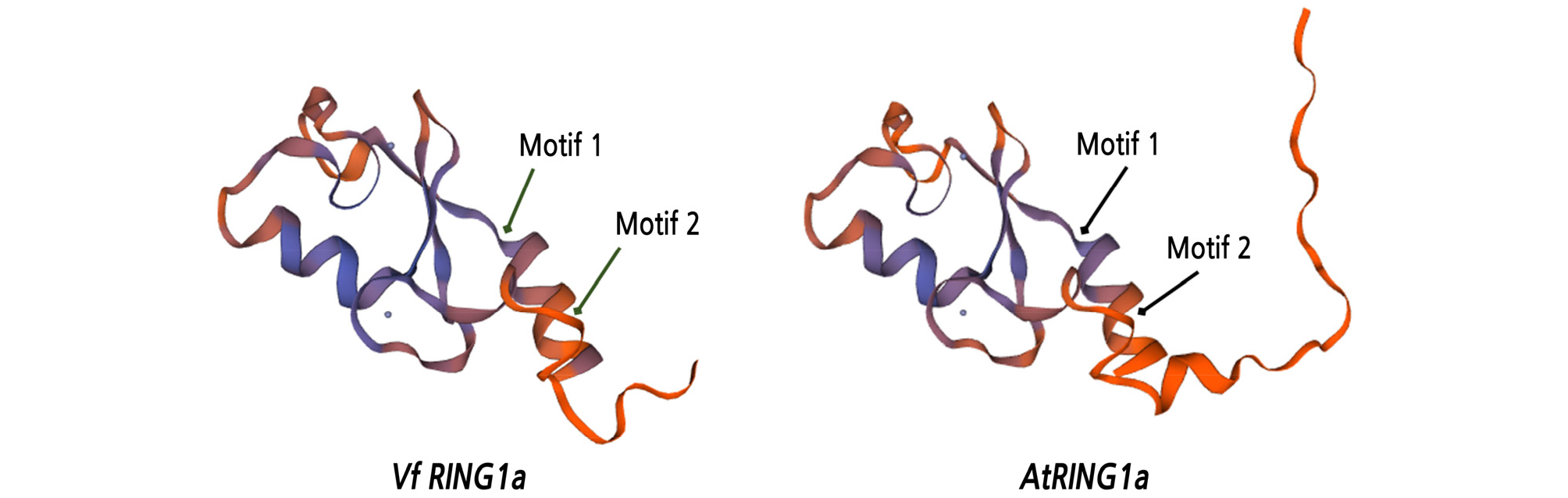
Three-dimensional models were built using a SWISS-MODEL protein structure prediction system, based on the predicted amino acid sequences of VfRING1a and AtRING1a. A model of the predicted ubiquitin ligase protein RING2, with motifs 1 and 2 of VfRING1a and AtRING1a, is shown in Fig. 3.
Expression Pattern of VfRING1a
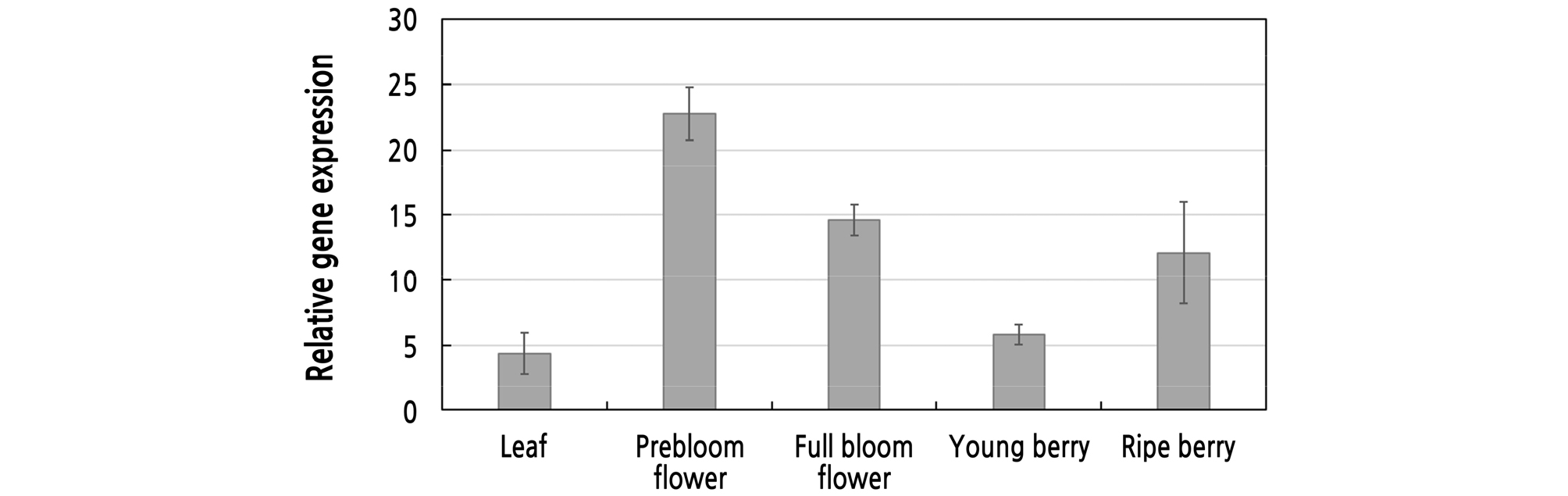
To investigate whether VfRING1a is expressed in different plant tissues, real-time PCR analysis was used to detect the VfRING1a transcription levels in varied tissues. VfRING1a expression was induced in leaves, in flowers in prebloom and full bloom stages, and in young and ripe berries of V. flexuosa (Fig. 4). We found that the VfRING1a gene is broadly expressed in multiple grapevine tissues. VfRING1a expression was high in prebloom and full bloom stage flowers, compared to the relatively low expression in vine leaves. This indicates that VfRING1a may be associated with flower development and blooming in grapevine.
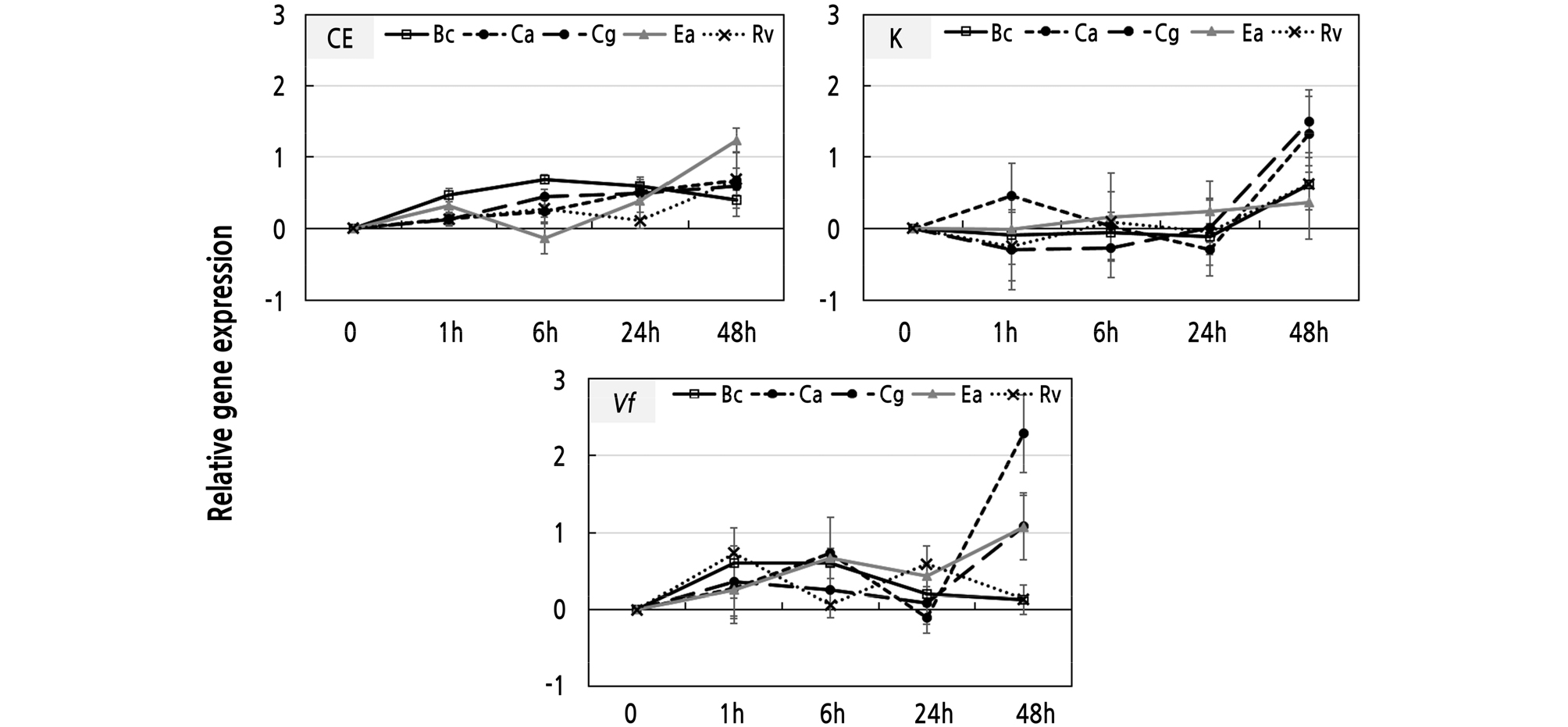
To investigate the VfRING1a expression levels induced during infection by pathogens, ‘Campbell Early’, ‘Kyoho’, and V. flexuosa grapevine leaves were inoculated with B. cinerea, C. acutatum, C. gloeosporioides, E. ampelina, and R. vitis. Our results show that VfRING1a expression is differentially induced and dependent on the infecting pathogen (Fig. 5). In ‘Campbell Early’, the B. cinerea-inoculated leaves induce the expression of VfRING1a at 6 h after inoculation and then it declines at 24 h (Fig. 5). VfRING1a gene expression in E. ampelina-inoculated leaves is induced at 1 h after inoculation, declines at 6 h, and increases at 24 h. In Colletotrichum spp.- and R. vitis-inoculated leaves, expression was induced at 1 h after inoculation of pathogens. VfRING1a gene expression in leaves inoculated with B. cinerea, C. gloeosporioides, E. ampelina, and R. vitis was initially down-regulated, with subsequent peaking of the expression at 48 h after inoculation after inoculation in ‘Kyoho’ grapevine (Fig. 5). Expression of VfRING1a in C. acutatum-inoculated leaves was induced at 1 h after inoculation, declined at 24 h, and then increased at 48 h. VfRING1a expression in B. cinerea-, C. gloeosporioides-, and R. vitis-inoculated leaves was induced at 1 h after inoculation, declined at 6 h, and increased at 24 h; in V. flexuosa grapevine, the expression was observed to subsequently decrease after 24 h (Fig. 5). VfRING1a expression in C. acutatum- and E. ampelina-inoculated leaveswas induced at 6 h after inoculation, declined at 24 h, and subsequently increased at 48 h.
Discussion
In this study, the VfRING1a gene was identified through structure characterization and RING domain analysis and was found to be similar to AtRING1a. Xu and Shen (2008) reported AtRING1a and AtRING1b as homologues of the Drosophila PRC1 subunit RING1. Previous studies have reported that the PRC1 complex relies on RING-finger proteins that regulate the chromatin structure by catalyzing the deposition of monoubiquitin on H2A to maintain transcriptional repression of genes (Molitor and Shen, 2013). The PRC1 complex is characterized by a conserved combination of two domains: an N-terminal RING domain and a poorly characterized C-terminal ubiquitin-like domain (RAWUL; RING finger- and WD40-associated ubiquitin-like) (Sanchez-Pulido et al., 2008). In this study, we found a RING-finger domain at the N terminus and a ubiquitin-like domain (RAWUL) at the C terminus in VfRING1a (Fig. 1). Feng et al. (2019) have also reported that there are two conserved domains in VvRING1a. Chen et al. (2010) reported that the AtRING1 and AtBMI1 proteins have nonredundant functions within a common PRC1-like complex involved in the repression of regulatory gene transcription and embryonic traits for proper somatic growth. In this study, a conserved protein domain was identified in VfRING1a, which is similar to that of AtRING1a. Therefore, it has been concluded that VfRING1a functions as a PRC1-like complex (similar to AtRING1a) in grapevines.
As shown in Fig. 1, the RING domain of VfRING1a contains eight conserved Cys and/or His residues, along with two zinc ions. These cysteine-rich RING-type domains, belonging to the conserved C3HC4-type RING-HC group, are known to be essential for catalyzing E3 ubiquitin ligase activity in the ubiquitin/26S proteasome system (Lovering et al., 1993; Lorick et al., 1999; Stone et al., 2005; Lin et al., 2008). This suggests that these protein sequences may play a role in plant defense response by modifying the proteome via ubiquitination. In plants, the RING domain of RING-finger proteins is specifically induced by pathogens, playing an essential role in plant defense responses (Zeng et al., 2006). In Arabidopsis, the RING-finger-type E3 ligases ARABIDOPSIS TOXICOS EN LEVADURA2 (ATL2) and ATL6 were rapidly induced in response to an elicitor (Salinas-Mondragón et al., 1999; Serrano and Guzman, 2004). The expression of rice EL5 and Arabidopsis BRH1 RING-type E3 ligase, which has in vitro ubiquitination activity, was induced by fungal elicitor treatment (Molnar et al., 2002; Takai et al., 2002). Therefore, the induction of VfRING1a by pathogen inoculation indicates that it might be associated with the response to biotic stresses in grapevines.
In this study, the expression level of VfRING1a in flowers in prebloom and full bloom stages was higher than that in other organs, indicating that VfRING1a possibly has an important function in floral organ development and blooming in grapevines. Similar to our findings, a previous study reported the induction of CaRING1 transcripts in flowers, although they were not induced in different plant tissues (Lee et al., 2011). It has been reported that PRC1 RING-domain proteins play a key role in floral development, such as in floral transition, floral organ development, and specification under PcG regulation (Calonje et al., 2008; Holec and Berger, 2012; Shen et al., 2014). Shen et al. (2014) also reported that AtRING1a acts as an important regulator of flowering in Arabidopsis, revealing an unknown role of a PRC1 RING-finger proteins in mediating the transition from vegetative to reproductive development in plants.
In conclusion, this study characterized VfRING1a, which contains a conserved RING-finger protein, in V. flexuosa grapevine. The conserved RING domain of VfRING1a is considered to play a role in transcriptional regulation and plant defense responses in grapevines. We further determined that the VfRING1a gene is differentially expressed in response to several pathogen infections in ‘Campbell Early’, ‘Kyoho’, and V. flexuosa grapevine leaves. Our results provide meaningful data for further study of the structure and function of the VfRING1a gene in development and defense responses in grapevines.