Introduction
Materials and Methods
Plant Materials and Cold Treatment
RNA Isolation and Real-time PCR Analysis
Cloning of 2OG Genes and Sequence Analysis
Comparison of Gene Structure
Results and Discussion
Introduction
Temperature is one of primary environmental factors that limit the growth, development, and productivity of crop plants. Molecular and genomic analyses have identified several regulatory systems controlling expression of genes in response to stress (Shinozaki et al., 2003). Grape (Vitis spp.), which is one of the most widely grown fruit crops in the world, is also influenced by low temperature during the growing season (Nam et al., 2012; Park et al., 2000; Warmund et al., 2008). Tattersall et al. (2007) Korean Journal of Horticultural Science & Technology 47 conducted a microarray analysis of V. vinifera cv. Cabernet Sauvignon that found that chilling affects many calcium signaling-related transcripts in shoot tips of grapevines exposed to chilling.
2-Oxoglutarate (2OG) is a molecule derived from the Krebs cycle, which is a highly conserved metabolic process. This compound plays a critical role in secondary metabolism as a signaling molecule in a variety of organisms (Zhao et al., 2010). 2OG and Fe(II)-dependent oxygenases (2OGO) are the largest known family of non-heme oxidizing enzymes, which have an absolute requireme nt for desaturation, hydroxylation, and oxidative ring closure reactions (Prescott and John, 1996). The 2OG and Fe(II)-dependent dioxygenases (2OGD) are the second largest family of plant enzymes and are also widespread in fungi, mammals, and microorganisms. 2OGDs also play roles in various oxygenation/hydroxylation reactions (Kawai et al., 2014). The 2OGD superfamily has been shown to participate in antibiotic biosynthesis, collagen biosynthesis, DNA repair, and stress responses (Aravind and Koonin, 2001).
The 2OGOs play a wide variety of biological roles in bacteria, plants, and human (Welford et al., 2005). 2OGOs function in adventitious root formation of apple trees (Butler and Gallagher, 1999), mugineic acid synthesis in barley (Okumura et al., 1994), and fruit ripening in tomato (Trentmann and Kende, 1995). 2OGs have been reported to regulate metabolic activities such as lipid absorption/metabolism, muscle performance, and cancerogenesis in the human body (Harrison and Pierzynowski, 2008), as well as nitrogen metabolism in Rhodospirillum rubrum (Teixeira et al., 2010). Cheng et al. (2014) also reported the function of four 2OG dependent oxygenases, anthocyanidin synthase (ANS), flavonol synthase (FLS), flavone synthase I (FNS I), and flavanone 3-β-hydroxylase (FHT), in plant flavonoid biosynthesis, as well as the relationship between their catalytic activity and tertiary structure in Apiaceae species. Finally, Araujo et al. (2012) identified the effects of 2OG on 2OGD gene expression in Arabidopsis leaves.
In a previous study, 34 2-oxoglutarate oxygenase (2OGO) transcripts with different expression levels were obtained from transcriptome data of V. labruscana treated with low temperatures (Ahn et al., 2014). We also identified six 2OGO genes in V. labruscana by comparing structures, nucleotide sequences, and deduced amino acid sequences. In addition, we analyzed the differential expression patterns in response to exposure to low temperature for 1–4 weeks in ‘Campbell Early’ grapevines.
Materials and Methods
Plant Materials and Cold Treatment
Grapevines of ‘Campbell Early’ with green shoots grown in a greenhouse were maintained at 4°C in a growth chamber under a 16:8 photoperiod for 0, 1, 2, 3, and 4 weeks. Healthy green leaves of grapevines were harvested at the indicated times, immediately frozen in liquid nitrogen and stored at -80°C for further analysis.
RNA Isolation and Real-time PCR Analysis
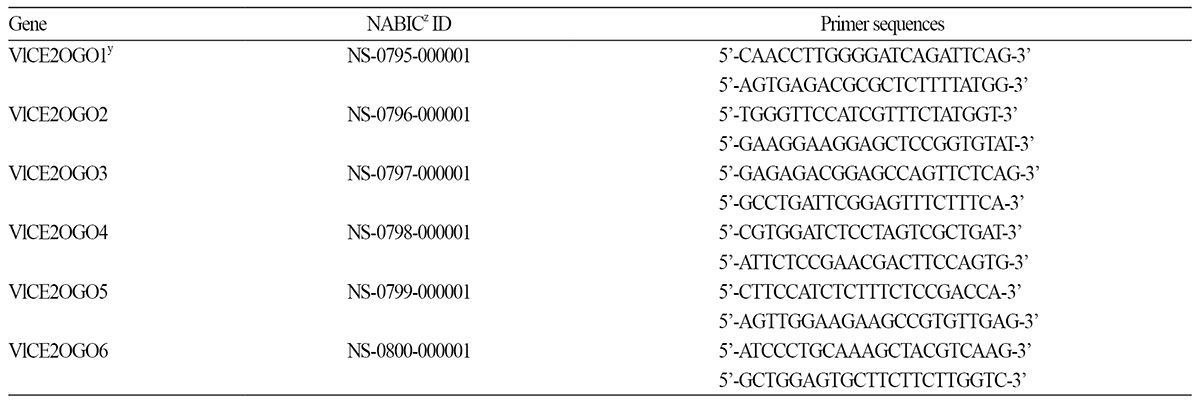
Total RNA was extracted from grapevine leaves harvested at each time point using the method described by Chang et al. (1993). First-strand cDNA was synthesized from the total RNA (500 ng) using a GoScript Reverse Transcriptome System (Promega, Madison, WI, USA). Real time PCR was conducted with a C1000TM Thermal Cycler (BioRad, Hercules, CA, USA) using SYBR Premix Ex (Takara Bio Inc., Osaka, Japan) as the fluorescent dye, then analyzed using the CFX Manager software (BioRad). The PCR amplification was conducted by initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Transcripts levels were normalized using the standard-curve method and calculated against the grapevine Actin gene (AB372563) as an internal control (Kim et al., 2014). The specific primer sequences of six 2OGO genes were designed by alignment of nucleotide sequences using Primer 3 (http://frodo.wi.mit.edu/primer3) for real-time PCR (Table 1), and used to investigate their expression in response to low temperature in grapevines.
Cloning of 2OG Genes and Sequence Analysis
Among 34 DEGs identified from transcripts of ‘Campbell Early’ kept at 4°C, by next generation sequencing (NGS), six loci showing different transcription levels with more than 100 scores were selected to characterize their structures and to evaluate their expressions against chilling treatment. Nucleotide and amino acid sequence data of VlCE2OGO1- VlCE2OGO6 (Vitis labruscana 2-oxoglutarate (2OG) and Fe(II) oxygenase 1-6) were deposited in the National Agricultural Biotechnology Information Center (NABIC), Rural Development Administration Korea database under accession numbers NS-0795-000001-NS-0800-000001 (Table 1), respectively.
Comparison of Gene Structure
Amino acid sequences were deduced from nucleotide sequences using the Translate program hosted by the ExPASy Proteomics Server (http://web.expasy.org/translate/). The primary structure of genes was performed using ProtParam (http://web.expasy.org/protparam/), while the secondary structure was analyzed using the Self-Optimized Prediction Method with Alignment (SOPMA) (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html.). The protein domain was analyzed using the Simple Modular Architecture Research Tool program (SMART) (http://smart.emblheidelberg. de/) (Letunic et al., 2014). Multiple alignment and pairwise comparisons of amino acid sequences were performed using T-coffee (http://tcoffee.vital-it.ch/apps/tcoffee/do:regular) (Notredame et al., 2000). A phylogenetic tree was constructed by the neighbor-joining method using MEGA version 6.0 (The Biodesign Institute, Tempe, USA).
Results and Discussion
Among six transcripts from ‘Campbell Early’ grapevine (V. labruscana), the highest similarity (38.6%) was observed between VlCE2OGO1 and VlCE2OGO2, while the lowest similarity (19.2%) was between VlCE2OGO2 and VlCE2OGO3 based on deduced amino acid sequences (Table 2). The deduced amino acid sequences of VlCE2OGO2, VlCE2OGO1, VlCE2OGO6, and VlCE2OGO4 showed 34.0, 33.7, 30.1, and 29.3% sequence similarity with those of VlCE2OGO4, VlCE2OGO4, VlCE2OGO2, and VlCE2OGO6, respectively.
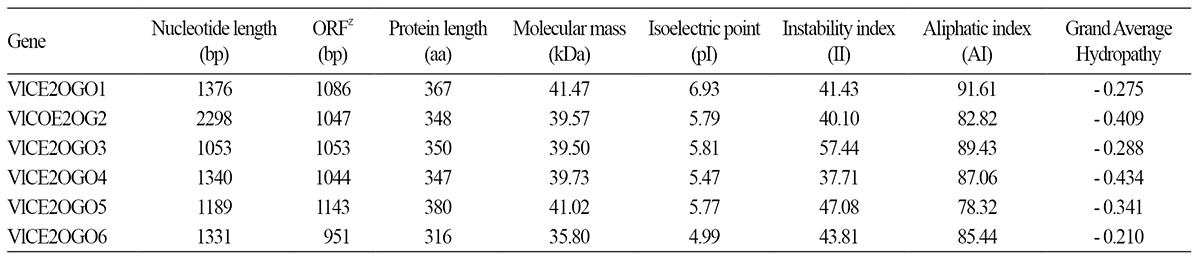
The primary structures of the six VlCE2OGO genes of ‘Campbell Early’ were characterized (Table 3). The six VlCE2OGO transcripts ranged from 2298 to 1053 bp in size. The sizes of the 2OGO genes varied in position and the sizes of the open reading frame (ORF) ranged from 92 to 1177, 57 to 1103, 1 to 1052, 109 to 1152, 47 to 1188, and 52 to 1002 nucleotides, respectively. The results showed that the size of the six 2OGO proteins varied from 316 to 380 amino acids (35.80 to 41.02 kDa). VlCE2OGO4 was identified as stable (II < 40) based on the instability index (II) proposed by Guruprasad et al. (1990).
The aliphatic index (AI) of a protein is a measure of the relative content occupied by the aliphatic residue side chain (alanine, valine, leucine, and isoleucine) (Gasteiger et al., 2005). AI index of the VlCE2OGOs varied from 78.32 to 91.61, which is regarded as a positive factor for the increase in thermal stability of globular proteins in grapevines. The grand average hydropathy (GRAVY) value for a protein is calculated as the sum of hydropathy values of all the amino acids divided by the number of residues in the sequence (Kyte and Doolittle, 1982). The GRAVY index of VlCE2OGO proteins ranges from -0.434 to -2.210, indicating that they are hydrophilic throughout the length of the protein.
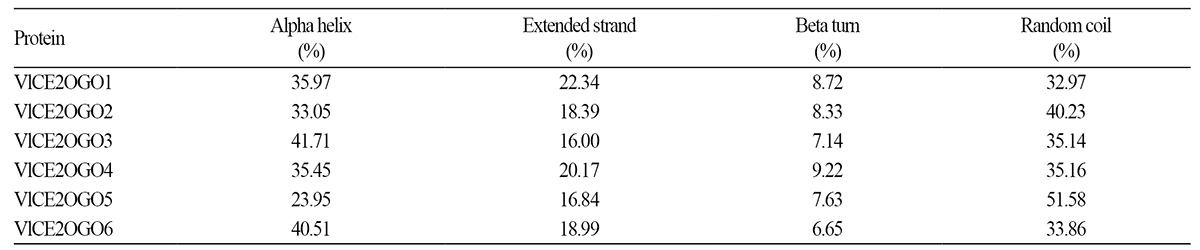
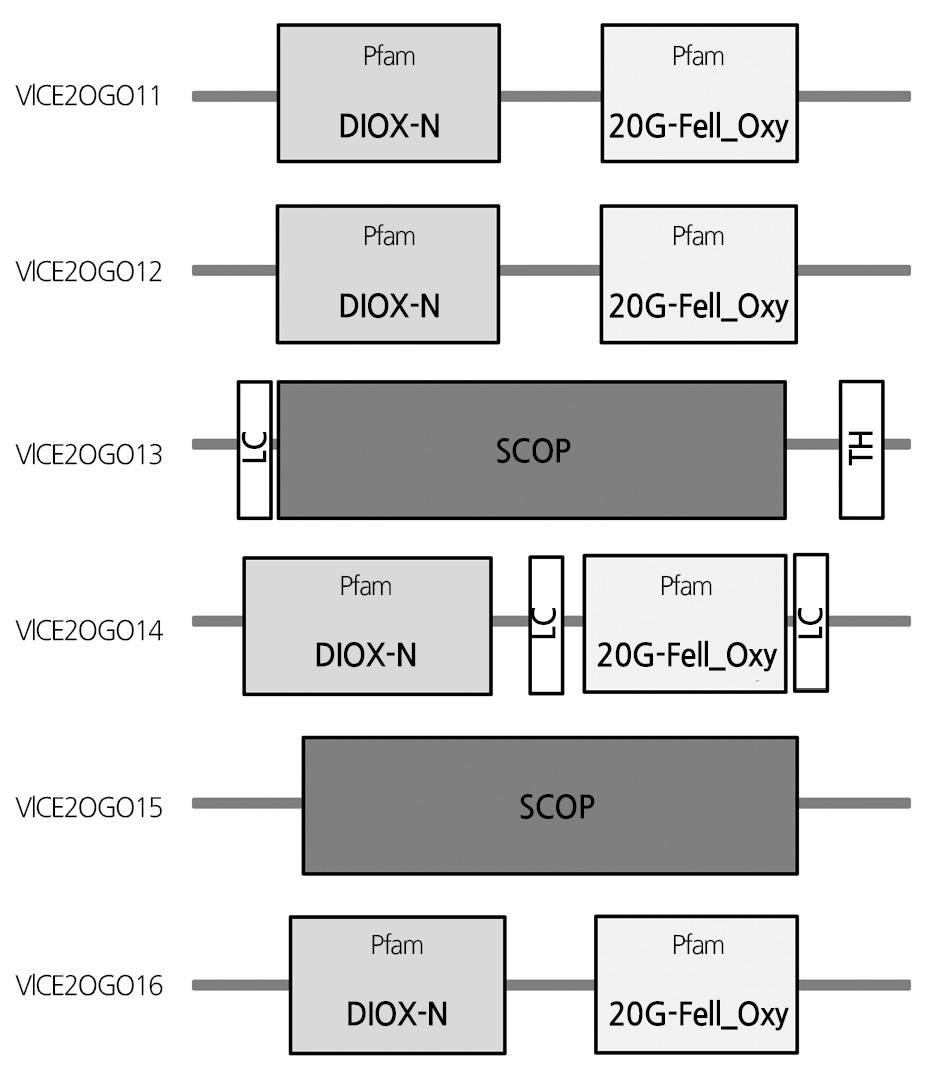
Secondary structure prediction of these proteins was conducted using SOPMA (Table 4). The results showed that alpha helixes, extended strands, beta turns, and random coils varied from 23.95 to 41.71%, 16.00 to 22.34%, 6.65 to 9.22%, and 32.97 to 51.58 %, respectively. Analysis of the structure of predicted proteins showed that most proteins have Pfam domains, including non-haem dioxygenase in the morphine synthesis N-terminal (DIOX_N) and 2OG-Fe(II) oxygenase superfamily (2OG-Fe-Oxy), which are conserved and essential domains of 2OGOs (Fig. 1).
Table 2. Identity of amino acid sequences of 2-oxoglutarate oxygenase (2OGO) proteins of Vitis labruscana.
| |
zVlCE2OGO: 2OGO proteins of Vitis labruscana cv ‘Campbell Early’. | |
Table 3. Primary structure analysis of 2-oxoglutarate oxygenase (2OGO) proteins of Vitis labruscana.
| |
zORF: open reading frame. | |
Table 4. Secondary structure prediction of 2-oxoglutarate oxygenase (2OGO) proteins of Vitis labruscana by SOPMA.
| |
Transmembrane helix (TH) was only detected in VlCE2OGO3. VlCE2OGO3 and VICE2OGO5 contained the structural classification of proteins (SCOP) domain. Multiple alignments of the amino acid sequences deduced from the nucleotide sequences of six 2OGO genes in the ‘Campbell Early’ grapevine are shown in Fig. 2.
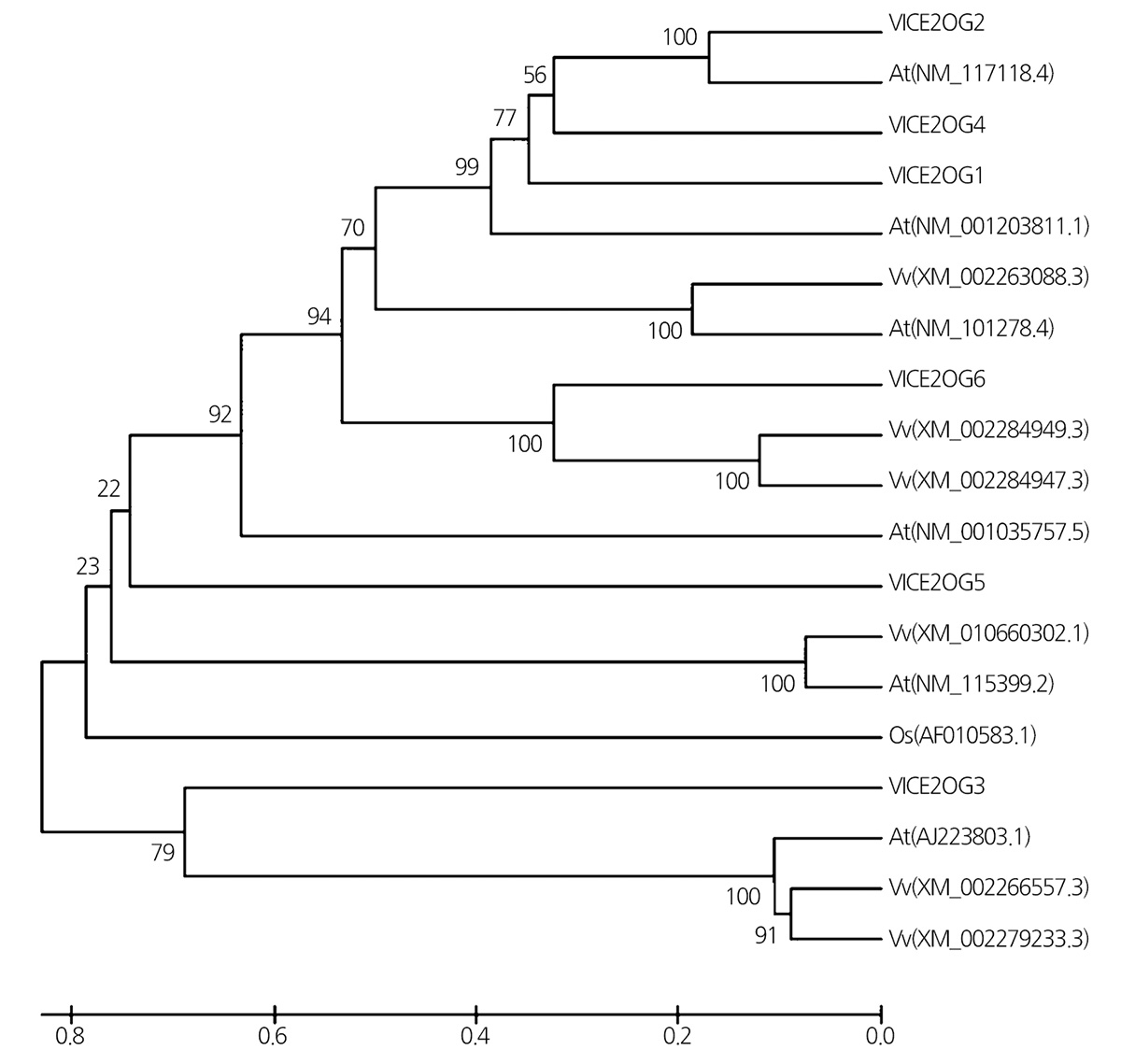
A phylogenetic tree based on the amino acid sequences of 2OGO transcripts was constructed to investigate the genetic relationships of VlCE2OGOs with other plant proteins (Fig. 3). The 2OGO transcripts were divided into two groups, one including VlCE2OG3 and another group comprising the remaining transcripts. Among VlCE2OGOs, the sequence of VlCE2OGO2 showed a high level of similarity to At2OGO (NM_117118.4).
| |
Fig. 2. Multiple alignment of amino acid sequences of unique regions from 2-oxoglutarate oxygenases (2OGOs). VlCE2OGO: 2OGO proteins of Vitis labruscana cv. ‘Campbell Early’. | |
| |
Fig. 3. Phylogenetic tree of 2-oxoglutarate oxygenase (VlCE2OGO) proteins from Vitis labruscana transcripts with 2OGO proteins from Vitis vinifera (Vv), Arabidopsis (At), and rice (Os). | |
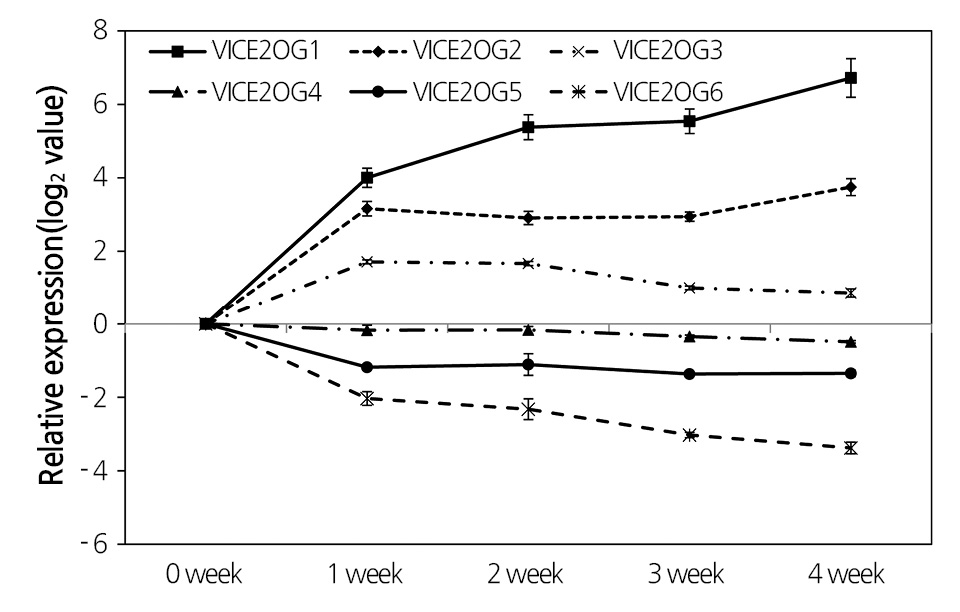
Expression of genes was determined by real-time PCR of grapevine leaves treated with low temperature using specific primers for each VlCE2OGO gene (Fig. 4). VlCE2OGO1, VlCE2OGO2, and VlCE2OGO3 genes were up-regulated, whereas VlCE2OGO4, VlCE2OGO5, and VlCE2OGO6 genes were down-regulated compared to controls in grapevines subjected to low temperature. Differential expression patterns from the real-time PCR analysis were consistent with those observed upon transcript analysis of grapevines maintained at low temperature for 4 weeks.
Low temperature is a serious abiotic stress that causes physiological damage and reduced productivity in growing grapevines. In the present study, we identified structural characteristics and analyzed the expression of six 2OGOs of ‘Campbell Early’ grapevine leaves in response to low temperature. The differential expression pattern of six 2OGOs was detected by real time PCR analysis with gene specific primers in ‘Campbell Early’ grapevines exposed to chilling temperature. Kim et al. (2014) reported that six basic helix-loop-helix (bHLH) genes showed up- and down-regulated expression patterns in response to low temperature treatments in ‘Campbell Early’ and ‘Muscat Bailey A’ grapevine leaves and latent buds.
2OG is related to the biosynthesis and secondary metabolism of signaling molecules such as alkaloids, gibberellins, and biosynthesis of flavonoids. Kawai et al. (2014) reported that 2OG Fe(II) dependent oxygenases were extensively present in various plant species. In Arabidopsis, more than 100 members of 2OGOs have been identified, which is greater than the number of identified and classified 2OGOs based on sequence analysis in a previous report (Presscot and Lloyd, 2000). Phylogenetic analysis showed that the six VlCE2OGO and other 2OGO genes were distributed in two major groups within several clades in the present study.
It has been reported that 2OGO was down-regulated throughout the cold treatment period and encoded a protein that demonstrated 44% identity with that of rice (Ciannamea et al., 2006). A hydropathy plot revealed that the predicted OsLti (Oryza sativa low-temperature-induced gene) proteins, OsLti6a and OsLti6b, were highly hydrophobic throughout the length of the protein, with GRAVY values of 1.51 and 1.42, respectively (Morsy et al., 2005).
Detection of 2OGD sequences has been reported in various plants by searching for the 2OG-Fe(II)-Oxy motif against whole amino acid sequences, with 142, 130, 114, and 74 2OGDs reported in the Picea, Arabidopsis, Oryza, and Selaginella genomes, respectively (Kawai et al., 2014). The presence of typical domain positions at 38–499 of the 2OG-Fe(II) oxygenase superfamily was detected in tea plants by ExPASy, BLAST, and SMART analyses (Rani et al., 2012). The flexibility and stability of 2OGO proteins have been influenced by dehydration and protein structure at random coils in response to water stress (Buxbaum, 2007; Qu et al., 1998). Schofield and Zhang (1999) suggested that biasing the reaction pathway of 2OGdependent oxygenases to deliberately generate radicals might lead to inhibition. A 2-OGD superfamily protein containing a 2OG-Fe(II)-oxygenase domain shared approximately 67% amino acid identity with downy mildew resistant 6 (DMR6) in Arabidopsis, and expression of DMR6 induced the expression of a subset of defense-associated genes such as pathogenesis-related genes against foreign stress (Damme et al., 2008). 2OGs were shown to have an activity of flavanone 3beta-hydroxylase (F3H) in ethylene synthesis and the biosynthesis of flavanoids, cathechins and anthocyanidins associated with defense responses to stress in plants including wheat (Giovanini et al., 2006) and petunia (Lukacin and Britsch, 1997).
In this study, we analyzed the structure, nucleotide sequences and deduced amino acid sequences of six 2OGO genes selected by transcriptome analysis by NGS of ‘Campbell Early’ grapevine leaves exposed to low temperature. All tested VlCE2OGO genes showed differential expression in response to low temperature treatment in ‘Campbell Early’ grapevine leaves. It suggests that induction of VlCE2OGO genes in expression level were related with defense responses to low temperature in grapevines. Further studies on gene expression based on transcriptome analysis in various organs like root and could provide meaningful information regarding the response of grapevine to stress from low temperatures in the future. The information presented herein will contribute to elucidation of mechanisms of cold hardness and selection of valuable molecular genetic resources responding to low temperature in grapevines.